Proposal for a New Interdepartmental Graduate
Degree Program in Bioinformatics (M.S. and Ph.D.)
To be offered by the
Interdepartmental Degree Committee in Bioinformatics
University of California, Los
Angeles
Submitted by: Christopher Lee, Department of Chemistry
and Biochemistry
Parag Mallick, Department of Chemistry and
Biochemistry
Matteo Pellegrini, Department of Molecular, Cell, and
Developmental Biology
Eleazar Eskin, Department of Computer Science
January 30, 2007
Table of Contents
Section 1. Introduction........................................................................................................ 5
Section
1-1. Aims and objectives of the
program...................................................... 5
Section 1-2. Historical development of the field and
historical development of the departmental strength in the field........................................................................................................................... 6
Section 1-3. The timetable for development of the
program..................................... 9
1-3a. Specific timing................................................................................................... 9
1-3b. Consistency of enrollment projections
with the campus enrollment plan......... 10
1-3c. Reduction of enrollments in programs in
order to accommodate the proposed program, if any 10
Section 1-4. Relationship of the proposed program to
existing programs on campus and the Campus Academic Plan. If the program is not in the campus
academic plan, why is it important that it be begun now?.............................................................................................................................. 10
1-4a. Could the curriculum be offered just as
effectively within an existing structure (e.g., as a pathway with an existing
major program)?........................................................................................... 11
1-4b. Overlap between the proposed curriculum
and the curricula of other units on this campus 12
1-4c. Effect of the proposed program on
undergraduate programs offered by the sponsoring department(s) 12
Section 1-5. Interrelationship of the program with
other University of California programs 13
1-5a. Possibility of cooperation or
competition with other programs within the University 13
1-5b. Differences from other similar programs
within the University and other California institutions 13
1-5c. Letters of evaluation from chairs of
departments in related fields from the UC system and outside. 16
Section 1-6. Department or group which will
administer the program................. 16
Section 1-7. Plan for evaluation of the program by
the offering department and campuswide 16
Section 1-8. Evidence that the different
participating disciplines contribute to the total program in such a way that
the student cannot achieve necessary knowledge without substantial study in two
or more established departments............................................................................................. 16
Section 2.
Program............................................................................................................ 17
Section 2-1. Undergraduate preparation for admission......................................... 17
2-1a. Field examinations and/or other
pre-qualifying examinations.......................... 17
2-1b. Qualifying examinations—written
and/or oral................................................. 17
2-1c. Relationship of master’s and doctor’s
programs.............................................. 17
2-1d. Special preparation for careers in
teaching....................................................... 17
2-1e. Other admissions requirements........................................................................ 17
Section 2-2. Foreign language requirement.............................................................. 18
Section 2-3. Program of Study................................................................................... 18
2-3a. Specific fields of emphasis............................................................................... 18
2-3b. Plan(s): Masters I and/or II; Doctorate............................................................ 19
2-3c. Unit requirements............................................................................................. 19
2-3d. Required and recommended courses,
including teaching requirement............. 20
2-3e. Requirements for licensing or
certification....................................................... 20
Section 2-4. Field examinations—written and/or
oral............................................. 20
Section 2-5. Qualifying examinations—written
and/or oral................................... 20
Section 2-6. Thesis and/or dissertation expectations............................................... 20
Section 2-7. Final examination requirements........................................................... 21
Section 2-8. Special requirements over and above
Graduate Division minimum requirements 21
Section 2-9. Relationship of masters and doctoral
programs................................. 21
Section 2-10. Special preparation for careers in
teaching....................................... 21
Section 2-11. Student sample program for each year.............................................. 21
2-12. Normative time from matriculation for
degree (assuming student has no deficiencies and is enrolled full-time)....................................................................................................................... 22
2-12a. Normative lengths of time for
pre-candidacy and candidacy periods............. 22
2-12b. Other incentives to support expeditious
times-to-degree................................ 22
Section 3.
Projected Need.................................................................................................. 22
Section 3-1. Student demand for the program........................................................ 22
3-1a. Documentation of demand for program
with three to five years of enrollment and admissions statistics from this or
other institutions, or data on rate of student inquiries............................. 23
3-1b. Evidence that this demand will be
stable and long-lasting................................ 23
3-1c. For new programs that are extensions of
existing disciplines, enrollment statistics from related courses to demonstrate
demand................................................................................................... 24
3-1d. Statistics or other documentation of
need......................................................... 24
Section 3-2. Opportunities for placement of
graduates........................................... 24
3-2a. Placement records of other UC programs
in the field in recent years............... 24
3-2b. Demonstration of a strong market for
program graduates by listing recent job listings, employer surveys, assessments
of future job growth............................................................................... 24
Section 3-3. The importance of the new program to
the discipline........................ 27
Section 3-4. Ways in which the program will meet the
needs of society................ 28
Section 3-5. Relationship of the program to research
and/or professional interests of the faculty 28
3-5a. Core faculty...................................................................................................... 28
3-5b. Associate faculty.............................................................................................. 29
Section 3-6. Differentiation of the proposed program
from existing UC and California independent university programs, and from
similar programs proposed by other UC campuses 30
Section 4. Staff.................................................................................................................. 30
4-1a. List of program faculty, their ranks,
their highest degree and other professional qualifications, and a citation of
no more than five recent publications (abbreviated curricula vitae); Data
concerning faculty limited to that information pertinent to the committee’s
evaluation of faculty qualifications....... 30
4-1b. Comments from chairs of departments
with graduate programs and/or faculty closely related to or affected by the
proposed program........................................................................................... 30
4-1c. For participating faculty members
outside of the sponsoring department, copies of letters indicating their
interest in the program (critical for interdisciplinary programs).................................. 30
Section 4-2. Organizational Structure....................................................................... 31
Section 4.3 Sources of available fellowship and
assistantship support................. 32
Section 5.
Courses.............................................................................................................. 32
Section 5-2. Elective Bioinformatics Courses........................................................... 32
The following
supporting courses are offered in related fields: Statistics 100A (or equivalent preparation) is required as a
prerequisite for Chemistry CM260A.
CS 31 (or equivalent programming skills) is required for Chemistry
C260BL (Bioinformatics Algorithms Laboratory). The Program in Computing offers a range of courses (PIC
10ABC, PIC 20AB, PIC 60, PIC 110) that are also very useful for bioinformatics
students. The many departments of
the Medical School and Life Sciences offer a wide range of coursework on
biology that is highly relevant to bioinformatics students.................................................................. 35
Section 5-3. Program degree requirements................................................................ 35
Section 6.
Resource Requirements and Enrollment Plans.................................................... 35
Section 6-1. Methods for funding.............................................................................. 35
6-1a. FTE faculty...................................................................................................... 35
6-1b. Staff FTE......................................................................................................... 36
6-1c. Library acquisitions.......................................................................................... 36
6-1d. Computing costs.............................................................................................. 36
6-1e. Equipment—inventory of current
equipment and future needs........................ 37
6-1f. Space and other capital
facilities—inventory of current facilities and future requirements 37
6-1g. Other operating costs (technical and
administrative staff, supplies and expenses, lab maintenance and other
facilities) and description of current staffing levels and future requirements.............. 37
Section 6-2. Projected doctoral enrollments for the
first five years........................ 37
Section 7.
Graduate Student Support.................................................................................. 37
Section 7-1. Strategy for meeting support needs..................................................... 37
Section 7-2. Current availability of faculty grants
to support graduate students and funding trends in agencies expected to provide
future research or training grants............................. 39
Section 7-3. Other extramural resources likely to
provide graduate student support, or internal fellowships and other
institutional support made available to the program......... 39
Section 7-4. Campus fund-raising initiatives that
will contribute to support of graduate students 40
Section 7-5. Graduate student support table listing
maximum number of students projected and sources of support for the first six
years of the program...................................................... 40
Section 8.
Changes in Senate Regulations........................................................................... 41
Section 9.
Abstract............................................................................................................. 41
Section 10.
Departmental Commitment to Proposed Program............................................ 41
Tables
Table 1. Sample
Student Program for the First Year........................................................... 22
Table 2.
Enrollments of Bioinformatics Core Courses ....................................................... 26
Table 3. Present
and Proposed Courses for the Program .................................................... 32
Table 4.
Projected Doctoral Enrollments for the First Five Years........................................ 38
Table 5. Graduate
Student Support.................................................................................... 40
Appendices
Appendix A. Summary of Information Required by the
California Postsecondary Education Commission
Appendix B. Letters of Support from Comparable
California Programs
Appendix C. Brief Curricula Vitae of Program
Faculty
Appendix D. Letters of Support from UCLA
Departments
Appendix E. Letters of Interest from Participating
Faculty Members
Appendix F. Catalog Descriptions of All Required
and Recommended Courses
Appendix G. Program Degree Requirements
Section 1-1. Aims and objectives of the program
We propose to
create an Interdepartmental Program (IDP) in Bioinformatics composed of faculty
from 14 departments, so that it is possible for students to apply for and
receive graduate training in bioinformatics at UCLA. This will strengthen faculty research in the Divisions of
Life and Physical Sciences and the Schools of Medicine and Engineering, help
train the next generation of scientists in a field of rapidly growing
prominence, attract new sources of funding to the UCLA campus, aid faculty
recruitment and retention, and drive growth at UCLA.
Our main goals
are:
- To provide integrated graduate training for
biologists, computer scientists and mathematicians that will enable them
to invent new kinds of bioinformatics throughout their future careers.
- To create a strong, collaborative community of
bioinformatics research groups at UCLA that unites faculty from many
departments, and fosters leading-edge research in this highly
interdisciplinary field. An
excellent graduate program is a critical success factor for world-class
research and for attracting superb faculty.
- To make major contributions to bioinformatics
research, through the development of new methodologies, new analyses of
the human genome and other genomics data, and discoveries that directly
affect human health.
Bioinformatics
can be defined broadly as the study of the inherent structure of biological
information. Some of this inherent structure is very
obvious (e.g., codon usage biases that reveal protein coding regions), while
others are less obvious but still immediately fruitful (e.g., how
transcriptional regulatory sites give rise to “programs” of gene expression),
while others are profound long-term challenges (e.g., how the genome encodes
the capabilities of the human mind).
Bioinformatics is the marriage of biology and the information
sciences. Long term, this is a
profound intellectual project.
Fortunately, it is producing immediately valuable results now, e.g.:
- Statisticians have invented analyses of DNA
microchip results (expression measurements of all 30,000 human genes
simultaneously) that can distinguish different types of tumors with
dramatically different treatment requirements, which previously were hard
to differentiate clinically.
- Evolutionary biologists have developed
bioinformatics analyses of genome sequence data that reveal the precise
pathways by which dangerous pathogens (like HIV) evolve drug resistance,
and how to slow the evolution of multi-drug resistance.
- Computer scientists have created powerful new
ways for mapping brain functions automatically from standard imaging data.
Bioinformatics
is of central importance to biomedical research in the 21st century
(see Section 1-2 below), and to the economy of California. By training both Bioinformatics M.S.
and Bioinformatics Ph.D. scientists from a variety of backgrounds, the proposed
IDP will contribute directly to the skilled workforce that California’s biotechnology
and software companies require for success. The IDP’s research may also give rise to new technologies
and new companies. Indeed, many of
the faculty have already done so in the past.
The proposed
IDP will provide an academic home for bioinformatics at UCLA that will bring
many different efforts together for the first time. Examples of current bioinformatics research conducted by the
core faculty include:
- The analysis of gene and protein sequences to
reveal protein evolution and alternative splicing
- The development of computational approaches to
study and predict protein structure to further our understanding of
function
- The analysis of mass spectrometry data to, for
example, understand the connection between phosphorylation and cancer
- The development of computational methods to
utilize expression data to reverse engineer gene networks in order to more
completely model cellular biology
- The study of population genetics and its
connection to human disease
UCLA has
already established a strong record of bioinformatics research and graduate
training (see Section 1-2 below).
In 1999 the faculty established a graduate core curriculum in
bioinformatics, which has been offered continuously since that time (see
Section 3-1a), demonstrating the faculty’s commitment to collaborative teaching
and to long-term development of an integrated bioinformatics program. These initiatives have been recognized
by a large number of awards of multi-investigator Project and Training grants
in bioinformatics from NIH, NSF, DOE and other funding sources. These many disparate efforts need a
strong graduate program to make them cohesive, successful, and competitive in
the long term.
The
establishment of the Bioinformatics IDP will allow UCLA to overcome the
limitations of the current situation, in which no single program brings
together bioinformatics students.
Specifically, we expect to resolve these existing weaknesses:
- Prospective bioinformatics graduate students do
not know which program to apply to at UCLA, since neither departmental
graduate programs nor ACCESS specifically recruit and train them.
- Graduate students at UCLA who are conducting
bioinformatics research currently take diverse courses that do not
necessarily cover the core training in bioinformatics that we believe they
need and that we propose here.
- Relative to other UC schools that already have
established bioinformatics graduate programs (see Section 1-5 below), the
lack of such a program at UCLA places UCLA at a disadvantage in competing
for the top bioinformatics students, and this impacts the ability of our
faculty to obtain funding in this area.
The creation
of the Bioinformatics IDP at UCLA will allow us to overcome all of these
limitations.
Section 1-2.
Historical development of the field and historical development of the
departmental strength in the field
Intellectually,
the proposed IDP draws strength from one of the most fundamental trends now
visible in the life sciences. Over
the last fifty years, biology has had one major theme—the marriage of
biology and physical chemistry (symbolized by the omnipresence of “molecular biology” throughout the life sciences). Its ultimate expression—the Human
Genome Project—has now been completed, setting the stage for a new
era. Over the next fifty years,
the most important engine of discovery will be the marriage of biology with the
information sciences, notably computer science.
The reason for
this is the revolutionary technological change in the biological sciences. It has always been a truism that
biology is information. However,
in the past biologists lacked the experimental and computational tools to deal
with the true complexity of biology’s information dimension. Instead of being able to see and
analyze the whole “computer program” of a cell or organism, biologists were
limited to breaking the computer down to its smallest components and measuring
their activity one gene at a time.
This is akin to trying to understand a computer’s program by attaching a
voltmeter to a single component inside it. While this reductionist program has been incredibly
productive in terms of discovering new components, it traditionally was often
difficult to put these pieces back together again in a way that could predict
or explain the behavior of the entire system.
Over the last
ten years, a wave of revolutionary technologies such as genomics has changed this situation dramatically. A marriage of molecular biology,
engineering and computer science has increased experimental bandwidth—the
ability to read out the program of information or activities in cells—by
up to 10,000 times what was possible before this automation. Where previously a research project
would sequence a gene, now it can sequence the entire genome. Where previously an experiment would
measure the expression of one gene, now it can easily and rapidly measure the
expression of all genes in the
genome. The dramatic completion of
the human genome five years before its planned deadline was only the most
visible example of this universal trend.
Already genome sequences of more than four hundred different organisms
have been completed. Many waves of
genomics technologies are spreading to new areas of biology, such as
proteomics, and this process is accelerating.
These
technologies have created an incredible avalanche of new experimental data, driving
the need for bioinformatics.
Genomics produces raw data; bioinformatics interprets its meaning.
Completion of the human genome, for example, led to a strange
anticlimax: once we had the data, it became obvious that we could understand very
little of what it means. The answers to this
problem—and there are already many—come from bioinformatics.
Already,
bioinformatics has gained enormous importance and prominence in the human
genome project and many other areas of biology, because of a simple shift in
the “rate-limiting step” of biological research. Previously, just getting experimental data was difficult and
slow. Now, biologists are awash in
huge amounts of experimental data (to cite just one example, over 400 fully
sequenced genomes); now the real challenge lies in analyzing their
meaning. This is bioinformatics.
A key feature
of this new field is its strongly interdisciplinary character. As we will document in Section 1-8,
there is no one department at UCLA that could conceivably cover the range of
topics—from Bayesian statistics to database design to protein
function—that come up in nearly every bioinformatics research
project. Statistics matter. Mathematical models matter. Computational algorithms and software
engineering matter. Genetics
matters. Molecular biology
matters. Protein structure
matters. There is no escaping the
problem of interdisciplinary training in bioinformatics. The proposed IDP would provide an
academic solution to this problem at UCLA for the first time.
UCLA has made
seminal contributions to bioinformatics research. UCLA’s strengths can be divided into several categories:
- Its bioinformatics faculty is strongly
interdisciplinary, spanning departments ranging from Statistics,
Chemistry, and Computer Science, to Human Genetics. UCLA is highly fortunate to bring
together all the capabilities of a world-class medical school, engineering
school, life sciences, mathematics and physical sciences, all in the
adjacent buildings of South Campus.
There are few universities that bring the full firepower of a
complete research university to bear on bioinformatics, as UCLA truly
does. The importance of the
fact that all of these are co-localized on one quadrangle—and many
meet every day for lunch at the café there—should not be
underestimated.
- The bioinformatics faculty is highly
collaborative. They have
established a six-year record of joint teaching of the bioinformatics core
curriculum, and a long list of multi-investigator collaborative research
projects, shared students, and joint training grants. The faculty has demonstrated that
they can work together to assemble successful joint research and training
programs.
- The faculty is strong in mathematical and
statistical approaches to bioinformatics problems, not only in the
Departments of Statistics and Mathematics, but also throughout the
group. Mathematical and
statistical approaches are widely recognized to be key to solving many
bioinformatics problems, and are of increasing importance in the field.
- The faculty combines experimental and theoretical
approaches integrally in their research. Whereas there is often a strong division between these
research perspectives, UCLA’s bioinformatics faculty work to integrate
theory and experimental analyses seamlessly, and have demonstrated that
they can do this very productively.
- The faculty is strong in both of the major
focuses of bioinformatics: genomics (focusing on DNA), and proteomics
(focusing on proteins).
Indeed, many of the faculty integrate both approaches to make discoveries
that would be impossible without this crossover.
- These diverse strengths have been recognized by
the selection of UCLA for establishment of a number of major centers in
bioinformatics, including the DOE- and Keck Foundation funded Centers for
Genomics and Proteomics; the first Integrated Graduate Education, Research
and Training (IGERT) grant in Bioinformatics funded by NSF in 2000; the
NIH-funded National Programs of Excellence in Biomedical Computing; the
NIH-funded Genomic Analysis training grant; and the NIH-funded Center for
Computational Biology.
Below we list
the faculty of the proposed IDP, indicating academic department (in bold),
bioinformatics IDP affinity group(s), and whether each is a core or an
associate member.
- James Bowie: Chemistry
& Biochemistry – structural biology (core).
- Guillaume Chanfreau: Chemistry
and Biochemistry –
genetics (associate).
- Ivo
Dinov: Statistics – neuroinformatics (associate).
- Joseph DiStefano: Computer
Science –
systems biology (core).
- David Eisenberg: Chemistry
& Biochemistry – structural biology (core).
- Eleazar
Eskin: Computer
Science –
genetics, computational methodology (core).
- Fred Fox: Microbiology,
Immunology & Molecular Genetics – bioinformatics in industry; industrial
contacts (associate).
- Tom Graeber:
Department
of Molecular & Medical Pharmacology – systems biology (core).
- Steve Horvath: Biostatistics – statistical methodology (core).
- Siavash Kurdistani: Biological
Chemistry –
genetics; systems biology (associate).
- James Lake: Molecular,
Cell & Developmental Biology – evolution (core).
- Ralf Landgraf: Biological
Chemistry – structural biology (associate).
- Ken Lange: Biomathematics – genetics; statistical methodology
(core).
- Christopher Lee: Chemistry
and Biochemistry – genetics; evolution; computational methodology
(core).
- Ker-Chau
Li: Statistics – statistical methodology (core).
- James Liao: Chemical
Engineering –
systems biology (core).
- Joseph Loo: Chemistry
and Biochemistry –
mass spectrometry (associate).
- Parag Mallick:
director of clinical proteomics at Cedars-Sinai Medical Center and
Chemistry and Biochemistry – mass spectrometry (core).
- Stan Nelson: Human
Genetics –
genetics (associate).
- John Novembre: Ecology
and Evolutionary Biology (Fall 2007) -- genetics (core).
- Stott
Parker:
Computer Science – computational methodology (core).
- Matteo
Pellegrini: Molecular,
Cell and Developmental Biology – systems biology; statistical
methodology (core).
- Peipei Ping: Physiology,
Medicine, and Cardiology – mass spectrometry (associate).
- Chiara Sabatti: Statistics – statistical methodology (core).
- Janet Sinsheimer: Human
Genetics –
statistical methodology; genetics; evolution (core).
- Stefano
Soatto: Computer
Science –
computational methodology; neuroinformatics (associate).
- Marc Suchard: Biomathematics – statistical methodology; genetics;
evolution (core).
- Charles Taylor: Ecology
and Evolutionary Biology – population genetics (associate).
- Arthur Toga: Neurology – neuroinformatics (associate).
- Yingnian
Wu: Statistics – statistical methodology (associate).
- Todd Yeates: Chemistry
and Biochemistry –
structural biology (core).
- Qing
Zhou: Statistics – developing statistical methods in cis-regulatory
analysis, including motif and cis-regulatory module discovery, gene
expression analysis, ChIP-chip data analysis, and comparative genomics
(core).
Section 1-3.
The timetable for development of the program
1-3a. Specific timing
Program approval: Because of substantial delays in the consideration of an
earlier proposal, we would like to have approval by July 2007, in order to
recruit a class for entry to the M.S. and Ph.D. programs in Fall 2008.
New faculty hires: Currently, the bioinformatics core faculty consists of 20
tenure-track FTE. Inevitably, this number will
grow as new hires are made in Bioinformatics. As evidence of the importance of this field at UCLA, five of
the 20 core faculty we list here have joined UCLA within the last year or will
join in 2007.
Course approvals: Chemistry CM260A was first approved in 1999. C260B was approved in Jan. 2007. 260BL, which is pending approval, was
previously part of the approved 260 course. Statistics M254 was approved in 1999. Chemistry 202 was approved in 1999.
First availability of core
offerings: The core courses for the
proposed IDP have been offered in most cases for several years: e.g., Chemistry
260 (Bioinformatics & Genomics), first offered Spring 2000; Chemistry 202,
first offered Fall 1999; Statistics M254, first offered Winter 2001; Chemistry
M252, first offered Spring 2002.
Availability of space/facilities
needed for program: Bioinformatics students
can currently access a variety of computational resources: (1) a room of desktop computers in the
Chemistry Building and (2) a cluster of computers in Boyer Hall.
Year of admission for first
cohort of graduate students: We
hope to admit the first cohort during the 2007-2008 application cycle, for
admission to study at UCLA beginning Fall 2008.
Anticipated year of awarding
first degrees: M.S. students admitted
Fall 2008 would graduate in 2010.
Ph.D. students admitted Fall 2008 would graduate most likely beginning
2013. It is possible that upon
approval of the IDP some existing UCLA graduate students would transfer into
the program to receive either the Bioinformatics M.S. or Ph.D. degree. (See list of current course enrollments
in Section 3-1c.) This could
advance the date of first degree awards to 2009-2010.
1-3b. Consistency of enrollment projections with the campus
enrollment plan
The planned
enrollments of the IDP (Table 1, page 23) are relatively small, and fit within
the campus enrollment plan, which calls for increased graduate enrollment in
line with supporting increased undergraduate enrollment (“Tidal Wave II”).
1-3c. Reduction of enrollments in programs in order to accommodate
the proposed program, if
any
We do not
anticipate a substantial reduction in enrollments in any existing program as a
result of the proposed IDP.
Currently, the largest category of students enrolled in the
bioinformatics core courses are admitted via the ACCESS program. Since the proposed IDP would create a
new degree area (UCLA has no existing bioinformatics Ph.D. or M.S. degree
program), it is reasonable to expect it would attract a new population of
students, rather than simply taking from existing admissions programs. However, even if we assume that every
ACCESS student enrolled in bioinformatics courses would apply to the proposed
IDP (and not ACCESS), the reduction to ACCESS would be slight (approximately
five students per year, which would be 6-9% of ACCESS yearly admissions of
55-75 students).
Section 1-4.
Relationship of the proposed program to existing programs on campus and
the Campus Academic Plan. If the program is
not in the campus academic plan, why is it important that it be begun now?
In 1999 the
School of Medicine conducted a strategic planning process, which identified
bioinformatics as one of the highest priorities for immediate development. One of the recommendations to emerge
from this review was for the deans to develop a specific plan for
bioinformatics training, which included a degree-granting education program.
The fact that
this recommendation was made five years ago indicates that the need for the
proposed IDP is urgent. It is
essential because it would bring together UCLA’s many academic resources in
bioinformatics for the first time.
The interdisciplinary character of UCLA’s bioinformatics
faculty—scattered among 11 departments in four schools—is both a
strength and a potential weakness.
As other prominent universities develop bioinformatics graduate degree
programs and competition grows, UCLA will be at a severe disadvantage in
recruiting top graduate student candidates if it does not offer a
bioinformatics degree and admissions program. First, UCLA does not offer a bioinformatics degree; instead,
students must apply to an existing department (e.g., Chemistry), and complete
its traditional degree requirements while performing dissertation research in
bioinformatics. For students who
are serious about bioinformatics research because they see it as a field of
great importance, not even having the word “bioinformatics” on their degree is
disadvantageous. Perhaps worst of
all, there is no way for an incoming student to access the full bioinformatics
faculty (who are in many different departments and schools). Effectively, students are forced to
guess who would be their dissertation advisor even before applying. Similarly, the scattered bioinformatics
faculty currently has relatively little voice in admissions decisions about
bioinformatics candidates, for the simple reason that there is no bioinformatics
admissions program.
Failure to
establish a graduate program would have negative effects on all of the existing
initiatives in bioinformatics and genomics at UCLA. Long-term, it is hard to imagine that the bioinformatics
training grant awards that UCLA has received would be renewed if the campus
failed even to begin a graduate program.
Faculty research in bioinformatics depends critically upon the quality
of graduate student recruitment.
Lack of a bioinformatics graduate program would become a major
disadvantage once other universities establish strong bioinformatics degree
programs, which is already happening.
This in turn would hurt the quality of bioinformatics research at UCLA,
and make it difficult to retain top bioinformatics faculty or recruit new
faculty.
1-4a. Could the curriculum be offered just as
effectively within an existing structure (e.g., as a pathway with an existing
major program)?
As noted in
Section 1-2, bioinformatics is a highly interdisciplinary field, and there is
no way that general bioinformatics graduate training can be offered in a single
existing UCLA department or program.
The potential problems would be numerous, including:
- Bioinformatics graduate students can (and should)
come from a wide variety of undergraduate backgrounds, including (but not
limited to) Biology, Biochemistry, Chemistry, Physics, Computer Science,
Statistics, and Mathematics.
There is no one existing department or program at UCLA whose
admissions requirements and criteria are appropriate for all of these
bioinformatics students.
- There is no existing UCLA admissions program that
includes all or even most of the bioinformatics faculty. Thus there is no program to which
students can apply that will allow them to do rotations and choose an
advisor from among the bioinformatics faculty. For example, while ACCESS includes many of the faculty
based in life sciences departments, it includes none of the faculty from
the engineering school, mathematics or statistics.
- Students who wish to focus on bioinformatics
often do not wish to be subjected to another extensive set of course
requirements (e.g., Chemistry) as a result of being forced to get their
degree from a traditional department. Because these requirements were established long before
the field of bioinformatics existed, many are not necessarily relevant to
bioinformatics study.
Meanwhile, there is much else to learn to become an expert in
bioinformatics.
- The interdisciplinary character of
bioinformatics—the fact that it has so many separate
pieces—creates a substantial danger that the student’s training will
fail to connect these pieces adequately. For example, it is essential to teach all the students
(be they from computer science, math or biology) a core of shared
vocabulary, concepts and methodologies, so that bioinformatics researchers
from different backgrounds can actually talk to each other, and work
together productively. Thus,
it is essential to create a new unified program where students learn
together and work together throughout their graduate training, to build
strong connections across their original disciplinary boundaries.
1-4b. Overlap between the proposed curriculum
and the curricula of other units on this campus
There is
relatively little overlap with existing programs at UCLA. Here we list all programs that have a nominal
overlap:
- ACCESS.
ACCESS is an umbrella graduate admissions program for the life
sciences at UCLA. It
organizes its faculty into a number of “affinity groups”, and has a
bioinformatics affinity group, consisting of several of the faculty from
this proposal who are located in life sciences departments. However, ACCESS neither provides
bioinformatics graduate training (its course requirements are designed for
traditional molecular biology/biochemistry research and have no
bioinformatics component), nor provides access to all of the
bioinformatics faculty at UCLA.
Many of the bioinformatics faculty are in departments not covered
by ACCESS.
- Biomathematics.
Biomath is a small department that emphasizes traditional
mathematical modeling of biological systems. With a few exceptions, its faculty do not perform
bioinformatics research, and its course requirements do not include
bioinformatics requirements.
- Biomedical Engineering. BME
is a relatively new graduate program in the School of Engineering that
emphasizes engineering approaches to biomedical problems, especially the
design of medical devices.
Its course requirements do not include bioinformatics courses, and
since most bioinformatics faculty are outside of the engineering school,
it does not include them.
- Human Genetics.
Human Genetics launched a graduate program about seven years
ago. Although the
department’s main focus is on experimental approaches (genetics and
genomics), bioinformatics is relevant to the interpretation of these
data. The department
inaugurated a “bioinformatics track” within its Ph.D. in Human Genetics,
to enable some of its students to gain credit for bioinformatics
coursework. However, the
Ph.D. course requirements overall remain focused on human genetics. Furthermore, most bioinformatics
faculty at UCLA are not in the Department of Human Genetics, and thus are
not included.
1-4c. Effect of the proposed program on
undergraduate programs offered by the sponsoring department(s)
Establishing a
strong graduate degree program in bioinformatics will be beneficial to
undergraduate programs in a number of departments. UCLA already offers both a number of undergraduate
bioinformatics courses (such as Chemistry C160A), and an undergraduate degree
(Computational and Systems Biology) designed for bioinformatics study. The growth and quality of these
curricula depend on the expertise of teaching assistants drawn from graduate
students in bioinformatics. Strong
undergraduate interest in the field of bioinformatics is demonstrated by
growing enrollments in courses such as Chemistry C160A, and in the
Bioinformatics specialization of the Computational and Systems Biology
major. Continued development of
undergraduate bioinformatics curricula requires a strong graduate program in
bioinformatics.
Section 1-5.
Interrelationship of the program with other University of California
programs
1-5a. Possibility of cooperation or competition with other programs
within the University
We do not see
significant competition with other graduate programs. As stated above, a few students may decide to apply to
the Bioinformatics IDP instead of ACCESS, but this would constitute only a
small fraction of the total number of students applying to ACCESS (about 9% at
most). Similarly a few of the students
who will apply to the Bioinformatics IDP would have applied to individual
departments (e.g., Chemistry) if the program were not available, but again this
would be a small number compared to the total number of students applying to
the department.
We see a
significant opportunity to collaborate with the ACCESS program in our
recruitment efforts. This could
also include cooperation in administering faculty presentations for the
students. The Center for
Computational Biology could provide administrative help in running the proposed
IDP, as graduate education is one of its priorities. Finally, we see an opportunity to leverage the computational
resources of the UCLA-DOE Institute for Genomics and Proteomics.
1-5b. Differences from other similar programs
within the University and other California institutions
Most of the
top research universities in California already have graduate degrees in
bioinformatics. Stanford, UC
Riverside, UCSF, UCSC and UCSD all offer Ph.D.s in Bioinformatics while UC Berkeley
and USC offer computational biology minors that accompany a traditional Ph.D.
in another program. Below we
describe these programs in detail and point to some of the similarities and
differences with the IDP proposed here.
UCSF
UCSF offers a
program in Biological and Medical Informatics (BMI) which encompasses data,
information, and knowledge acquisition, representation, modeling, integration,
communication, and interpretation ranging across basic science and engineering
through clinical practice and policy.
The primary mission of the BMI Program is to train biomedical
informatics researchers for academia and industry. The Program's focus is on the science of biomedical
informatics, with special emphasis on rigorous methodology, innovation, and generalizability
of findings, rather than the routine application of technology to biomedical
science and practice. Training
spans the full spectrum of biomedical informatics—from bench to bedside
to health system. Graduates of the
Program are well positioned to contribute at the interface of bio- and medical
informatics, where future research opportunities are excellent.
The Program
offers training leading to both the M.S. and the Ph.D. degrees. The M.S. program is open only to
students with a health sciences-related graduate or professional degree (e.g.,
M.D., Pharm.D., M.S. in epidemiology); the Ph.D. program is open to students
with a baccalaureate degree and the appropriate prerequisites.
All students
take the same curriculum in the first year. In the second year, students present their work in a
seminar, which is evaluated by selected faculty. Masters' students then work on a final research project,
while Ph.D. students prepare for their oral qualifying examination. Upon successful completion of their
orals, Ph.D. students advance to candidacy and begin work on their
dissertation. A thesis defense is
required for graduation.
The UCSF
program is more focused on medical informatics than the one we propose
here. Unlike our IDP, which
integrates expertise from medicine, engineering, life sciences, math and
physical sciences here, UCSF’s is based solely in a medical school.
UCSC
The Department
of Biomolecular Engineering offers B.S., M.S., and Ph.D. degrees in
bioinformatics. Bioinformatics
combines mathematics, science, and engineering to explore and understand
biological data from high-throughput experiments, such as genome sequencing and
gene expression chips. The program
builds on the research and academic strengths of the faculty in the Center for
Biomolecular Science & Engineering.
The immense
growth of biological information stored in computerized databases has led to a
critical need for people who can understand the languages, tools, and
techniques of mathematics, science, and engineering. A classically trained scientist may be unfamiliar with the
statistical and algorithmic knowledge required in this field. A classically trained engineer may be
unfamiliar with the chemistry and biology required in the field. This program strives for a balance of
the two; an engineer focused on the problems of the underlying science, or,
conversely, a scientist focused on the use of engineering tools for analysis
and discovery.
The graduate
degree prepares the student for life as a cutting-edge researcher in
bioinformatics, creating new tools to answer new questions. Support is available to graduate
students through an NIH training grant in bioinformatics, CBSE Diversity
Fellowships in Genomic Science, and various other sources detailed on the CBSE
website.
The primary
difference between our IDP and the UCSC Bioinformatics program is the breadth
of the core faculty, which in our case spans 11 departments, whereas the UCSC
program is more focused, offered by one academic department. Thus the programs’ scientific strengths
differ significantly in their mix of disciplines.
UCSD
The
Bioinformatics graduate program draws upon the interdisciplinary expertise of
affiliated faculty from seven participating departments and graduate
programs: Bioengineering, Biology,
Biomedical Sciences, Chemistry and Biochemistry, Computer Science and
Engineering, Mathematics, and Physics.
In addition, UCSD offers an interdisciplinary specialization in
Bioinformatics.
In recent
years, Bioinformatics has been identified by the UCSD administration as one of
the most important growth areas for the campus. Several recent new faculty hires have been targeted in
bioinformatics-related fields.
UCSD has also seen a significant increase in the research activity
associated with Bioinformatics across the traditional disciplines.
In general,
the UCSD program is very similar to the one we are proposing for UCLA and could
be used as a model.
USC
Computational
molecular biology has been thriving at USC since 1982, when Michael Waterman
joined the Departments of Mathematics and Biological Sciences. Since that time the group has grown to
include eight professors and numerous research students and postdoctoral
associates.
USC’s approach
to research and training in the Computational Biology and Bioinformatics area
has biology as its essential motivation.
An important aspect involves problem formulation, an interactive process
that involves collaborations between biologists and mathematical scientists. Once a problem is formulated, the solution
often involves a blend of statistics and algorithms. This is why the group has had close associations with three
departments: Mathematics, Biological Sciences, and Computer Science. Several of the faculty have joint
appointments in these departments, as well as in the Keck School of Medicine.
Students
wanting a Ph.D. in Applied Mathematics or Computer Science with an emphasis in
computational biology may enroll directly in the appropriate Ph.D.
programs. These students must
satisfy the relevant degree requirements of their home department, in addition
to a number of additional courses (chosen in consultation with faculty
advisors) that ensure appropriate breadth in biology, computer science and
mathematics. In Fall 2003 USC
began a Ph.D. program in Computational Biology and Bioinformatics, based in the
Department of Biological Sciences.
In contrast to
the IDP we are proposing for UCLA, the USC program is run out of a single
department. The emphasis in
computational biology that is offered at USC, which does grant students access
to faculty from three departments, differs from our proposal in that it
supplements the existing Ph.D. degrees offered by these departments, and
therefore requires substantially more coursework than the Ph.D. program we are
proposing.
Stanford
The mission of
the Stanford Biomedical Informatics (BMI) program is to train the next
generation of researchers in biomedical informatics. Specifically, their students develop and apply methods for
acquiring, representing, retrieving, and analyzing biomedical knowledge and
data. The BMI training program
encompasses bioinformatics, clinical informatics, and public health
informatics. Bioinformatics
focuses on methods for relevant to basic biology. Clinical informatics focuses
on methods relevant to patient care.
Public health informatics focuses on methods relevant to entire health
systems. Examples from these areas
can be found in the list of interests of their participating faculty.
As in the case
of UCSF, Stanford’s program in bioinformatics is focused on medical
applications and not basic science.
As seen in the breadth of research of our core faculty, our research
spans both medical informatics and basic science, and therefore offers graduate
students a much broader research base to choose from.
UC
Riverside
The Genetics,
Genomics and Bioinformatics (GGB) Graduate Program leads to conferral of a
Ph.D. in GGB, with a subspecialization in Genomics/Bioinformatics. The GGB Program also administers a full
undergraduate curriculum in Genomics/Bioinformatics within the Biological
Sciences major.
The program is
diverse, with research opportunities ranging from plant breeding to
population/evolutionary genetics to molecular biology to bioinformatics. Robust cross-disciplinary interactions
are fostered, using genetics as a common theme. These collaborations often bridge fundamental and applied
genetics that merge the basic and agricultural life sciences with statistics
and computer sciences. This same
intellectual diversity also offers incoming students a wide variety of choices
for conducting their dissertation research. Trainees are able to choose among one of the three available
curricular tracks. These tracks
have been created so that there is significant overlap in coursework. Personalized academic programs are
easily developed.
The Genomics & Bioinformatics track was implemented in the 2001-2002 academic year
and is rapidly becoming a very popular option within the Genetics Ph.D.
program. The track melds relevant
Statistics, Computer Science and Life Science courses into a flexible
curriculum that would appeal to computationally-oriented informaticians or to
"wet-bench" -oriented genomicists. This curricular track has helped the GP retain its unique
identity in an environment of rapid programmatic expansion in the College of
Natural and Agricultural Sciences.
The primary
difference between UC Riverside’s program and ours is that the Bioinformatics
component is part of a broader program in Genetics, Genomics and
Bioinformatics. We are proposing a
IDP that is exclusively focused on Bioinformatics in order to attract the top
students in this area.
UC Berkeley
The Graduate
Group established the Designated Emphasis (DE) in Computational and Genomic
Biology to provide specialized multi-disciplinary training and research
opportunities in the different facets of computational biology and
genomics. The DE is a
specialization offered by existing Ph.D. programs. At UC Berkeley, acquiring a DE is like earning a “minor”
with a Ph.D. degree.
In this sense,
the Berkeley program differs significantly from ours, which would grant a Ph.D.
in Bioinformatics, and not merely a minor. This should provide us a significant competitive advantage
in recruiting students who are interested in bioinformatics research.
1-5c. Letters
of evaluation from chairs of departments in related fields from the UC system
and outside.
See Appendices
D.
Section 1-6.
Department or group which will administer the program
The Bioinformatics IDP will report to the Dean of Life
Sciences and be administered by the IDP Graduate Program Committee. Program administration is described in
Section 4-2.
Section 1-7.
Plan for evaluation of the program by the offering department and
campuswide
The IDP will
first be reviewed after two years of operation, by an internal review committee
consisting of the Executive Committee and additional members appointed by the
Deans of the Schools of Medicine and Engineering and the Deans of the Divisions
of Life Sciences and Physical Sciences.
Results of the review will be provided to all four Deans. Subsequently, the IDP will undergo
external review every eight years.
We will also establish an external advisory board consisting of
distinguished scientists from industry.
Section 1-8.
Evidence that the different participating disciplines contribute to the
total program in such a way that the student cannot achieve necessary knowledge
without substantial study in two or more established departments
Bioinformatics
is the marriage of biology and the information sciences. As such it depends on substantial study
in both of these fields, which cannot be achieved without study in two or more
existing departments. This is
perhaps best illustrated by a typical example of a bioinformatics laboratory at
UCLA. Bioinformatics analyses in a
UCLA laboratory (Lee) have demonstrated the ubiquitous importance of
alternative splicing in human genes.
These analyses required thorough understanding of: the molecular biology
of mRNA processing; the experimental methods, data, and pitfalls of genomics
detection of mRNAs; computer science algorithms of sequence analysis and
alignment; statistical methods for measuring evidence such as Hidden Markov
Models, and hidden mixture deconvolution; computer database design and
data-mining technologies; software engineering techniques for massively
parallel computing. This collection
of skills spanning several disciplines is the rule, not the exception, in
bioinformatics.
Currently,
there is no UCLA graduate program that offers instruction in all of these
skills in one place.
Section 2. Program
Section 2-1.
Undergraduate preparation for admission
2-1a. Field
examinations and/or other pre-qualifying examinations
Applicants must take the
Graduate Record Examination (GRE) General Test, and a GRE Subject Advanced Test
in the area of the applicant’s undergraduate major is highly recommended. Since there is no GRE subject test in
Bioinformatics, such students should take a subject test in Biology,
Biochemistry, Computer Science or Mathematics.
2-1b. Qualifying examinations—written
and/or oral
There is no requirement for
an additional examination for admission to the IDP.
2-1c. Relationship of master’s and doctor’s
programs
Admissions requirements will
be identical for the M.S. and Ph.D. programs. Typical M.S. students will be primarily receiving Ph.D.
degrees from other campus degree-granting programs. Consequently, M.S. students will have the option of
transferring to the Ph.D. program subject to the approval of the program
administration. Similarly, Ph.D.
students transferring to alternate campus programs may receive the M.S. degree
upon completion of its requirements.
2-1d. Special preparation for careers in teaching
No special preparation for
teaching will be required for admission to the program.
2-1e. Other admissions requirements
Expected undergraduate
preparations for the program fall into three major categories:
- Bioinformatics and computational biology
majors: an increasing number of
universities are offering undergraduate majors in bioinformatics and
computational biology. (UCLA
itself offers undergraduate bioinformatics study via its Computational and
Systems Biology major). This
represents an ideal undergraduate preparation for the program, because it
demonstrates the applicant’s performance in each of the essential subject
areas—biology, mathematics, and computer science.
- Double majors in biology and computer science
or mathematics: in our view, the single greatest
difficulty of bioinformatics is its interdisciplinary character. Most undergraduates only have
strong preparation in one subject area, making it difficult to evaluate
their likely performance in other areas that are essential for
bioinformatics. We can
control for this risk by recruiting students who have double majored in a
life science plus a quantitative science, preferably computer science or
mathematics.
- Single majors with strong evidence of
interdisciplinary skills: in some cases we will admit
exceptionally strong students from a single traditional major. In this case, the student’s
academic record and research experiences must show clear evidence of
ability in other areas essential for bioinformatics, outside of his/her
major. In particular,
non-life science majors must demonstrate strong performance in relevant
life science coursework.
Similarly, life science majors must demonstrate strong quantitative
skills in computer science and mathematics.
It should be emphasized
that the program is seeking to build a small, high quality graduate program,
and thus intends to be highly selective. For example, we intend to concentrate
on students with double majors or (increasingly over time) computational
biology majors.
The UCLA Computational and
Systems Biology major with Specialization in Bioinformatics provides an example
of a template of appropriate undergraduate preparation for the program, albeit
with a strong engineering background (http://www.cs.ucla.edu/~cyber/major_majorfieldrequirements.htm).
Other undergraduate
preparations that are more life science-oriented are equally valid. During this period of rapid change in
bioinformatics and the early development of bioinformatics curricula, no fixed
formula for appropriate preparation can be enforced. Flexibility and case-by-case evaluation of a student’s
demonstrated skills and interests are essential.
Additionally, the program
will place a strong emphasis on applicants’ bioinformatics research
experience. Currently, most
students entering UCLA via various departments to pursue bioinformatics have
extensive bioinformatics research experience. Success in bioinformatics research, and strong letters of
recommendation from bioinformatics faculty advisors, provide the program with
clear evidence of a student’s ability to combine the interdisciplinary skills
necessary for bioinformatics. This
is very important, and will continue to be a vital consideration for
admissions.
As is standard for all UCLA
graduate programs, the Test of English as a Foreign Language (TOEFL) will be
required for admissions of students who are not native speakers of English.
Section 2-2.
Foreign language requirement
Ability in a foreign
language, while recommended, is not required for the bioinformatics
program. English is ubiquitous as
the language of bioinformatics journals, conferences and discussion, and is spoken
as a first or second language by nearly all researchers in the field.
Section 2-3.
Program of Study
2-3a. Specific fields of emphasis
We believe that in
bioinformatics, creative discoveries will come from "crossover"
collaboration between experts in different disciplines, and that special
training is required to give scientists the vocabulary and skills for this
teamwork. Over the years we have
developed and refined the following basic elements designed to address this
challenge:
- Bioinformatics Core Curriculum:
All students must take the core curriculum, designed to teach the
core concepts and methodologies which researchers must know to invent new
kinds of bioinformatics. In
other words, rather than just teaching examples of “bioinformatics past”
(as is the case for some bioinformatics courses), these courses define the
hard core of fundamental concepts—information theory, probabilistic
measures of evidence, and computational complexity—that underlie the
invention of each new kind of bioinformatics. Most importantly, the core curriculum gives students
from widely varying backgrounds a common vocabulary and set of concepts,
so they can communicate and collaborate with researchers throughout the
field of bioinformatics.
- We segment bioinformatics into a number of
existing application areas, which are likely to grow in the future:
- Genomics: focused on the
bioinformatics of genome biology.
Genomics examples include genome assembly, microarray gene
expression analysis, QTL and disease gene mapping, alternative splicing,
and comparative genomics.
- Proteomics: the bioinformatics
of protein structure and function.
Proteomics examples include protein interaction networks, protein
isoform identification by mass spectrometry, protein domain and structure
prediction.
- Neuroinformatics:
bioinformatics of neuronal systems, including three-dimensional
imaging of brain structures.
Examples include brain mapping, image analysis.
- Computer science:
focused on algorithms and architectures of bioinformatics computation. Examples include Hidden Markov
Models, bioinformatics database architecture, and massively parallel
algorithms.
- Math and Statistics:
focused on mathematical analysis and models of bioinformatics
problems. Examples include
Bayesian statistical methodologies for gene expression analysis,
statistical genetics, and bioinformatics applications of information
theory.
2-3b. Plan(s): Masters I and/or II; Doctorate
M.S. candidates may fulfill
the requirements of the degree either by writing a master’s dissertation or by
passing an oral examination. The
Ph.D. degree requires an oral qualifying exam and oral dissertation
defense. Following completion of
course requirements all students will be expected to take and pass a qualifying
exam that will test their proficiency in each of the concentration areas and
their ability to integrate the disparate disciplines.
2-3c. Unit requirements
Both M.S. and Ph.D. students
must complete the core curriculum (three four-unit lecture courses and two
three-unit lab courses), and an elective area (at least three four-unit courses
from a designated list). M.S.
students must also complete at least 12 units of laboratory research. Ph.D. students must complete an
additional elective course, at least 12 units of laboratory rotation (596), and
at least six units of seminar courses.
This constitutes a total of 42 units for the M.S. degree, and 64 units
for the Ph.D. degree.
2-3d. Required and recommended courses,
including teaching requirement
Both M.S. and Ph.D. students
must complete:
- The Core Curriculum: Bioinformatics and Genomics (Chemistry CM260A, formerly
Chemistry 260), Advanced Algorithms for Bioinformatics (Chemistry C260B, a
new course pending approval), Statistical Methods in Computational Biology
(Statistics M254, proposed to be cross-listed as Chemistry M260C),
Bioinformatics Algorithms Laboratory (Chemistry 260BL, a new course),
Bioinformatics Interdisciplinary Research Seminar (Chemistry 202, a
year-long course), Advanced Methods in Computational Biology (Chemistry
252, a year-long course). See
Section 5 for details.
- Elective courses from concentration areas. Each student’s course list will be
subject to approval by an IDP faculty advisor. See Section 5 for recommended course listings.
To help Bioinformatics IDP
students to develop their teaching skills, they will be required to do a
minimum of one quarter of teaching assistantship. Students will be encouraged to gain additional teaching
experience both for bioinformatics courses and courses in their major
area.
2-3e. Requirements for licensing or certification
There are no licensing or
certification requirements for the program.
Section 2-4.
Field examinations—written and/or oral
The program will not require
a field examination.
Section 2-5.
Qualifying examinations—written and/or oral
Ph.D. degree candidates, but
not M.S. candidates, will be required to pass a written qualifying examination,
consisting of a research proposal outside their dissertation research topic,
and an oral qualifying exam defending their dissertation research proposal
before their dissertation committee.
Moreover, before taking the Ph.D. qualifying exams, the student must
first complete the Core Curriculum.
Following qualifying exams and advancement to candidacy, students must
present a summary of their research to date and proposed future research within
an oral seminar format. This
summary should be presented approximately one year after advancement to
candidacy.
Section 2-6.
Thesis and/or dissertation expectations
M.S. degree candidates may
opt to write a dissertation to fulfill the requirements of the M.S.
degree. In this case, the
candidate should choose an IDP faculty advisor and submit a dissertation
proposal by the end of the third quarter of study. To proceed, the proposal must be approved by a faculty
member who agrees to become the student’s dissertation advisor. Completed dissertations will be
evaluated by a committee of at least three IDP faculty members, which must be
approved by the IDP Director. The
student must present the completed dissertation in a public seminar.
Ph.D. students must write a
dissertation. During their first
year, Ph.D. students will perform laboratory rotations with IDP faculty whose
research interests them, and must select a dissertation advisor from the IDP
faculty by the end of their third quarter. Next, students must select a qualifying committee by the end
of their second Spring quarter, and submit a written dissertation proposal by
the end of their second year. The
qualifying committee must consist of the faculty advisor and at least three
additional faculty, of whom two must be IDP faculty. The qualifying committee must be approved by the IDP
Director and the campus graduate Dean.
The written dissertation proposal must be given to all the qualifying
committee members prior to the scheduled oral qualifying exam. Students will advance to candidacy when
they pass the oral qualifying exam, complete the IDP course requirements, clear
all incompletes from their transcript, and pay the filing fee. Students should advance to candidacy by
the end of their third Fall quarter.
Failure to advance to candidacy by the end of the fourth year will
result in academic probation.
The completed dissertation
must be submitted to the reading committee three weeks before the dissertation
defense. The reading committee
must consist of the faculty advisor and at least two additional readers from
the IDP faculty, and must be approved by the IDP Director. The candidate must present a public
seminar, immediately followed by the dissertation defense. The committee must determine whether
the dissertation completely fulfills the requirements of original, important,
publishable research in bioinformatics, and whether revisions are required. Ph.D. dissertations must conform to a
high standard of originality, relevance and impact suitable for publication in
peer-reviewed journals in the field.
Successful completion of the dissertation and defense constitutes the
final requirement for the Ph.D. degree.
Section 2-7.
Final examination requirements
No written examinations are
required for the M.S. or Ph.D. degrees.
M.S. candidates may fulfill the requirements of the degree by passing an
oral examination. Ph.D. candidates
must pass both a qualifying examination and dissertation defense described
above in Section 2-6.
Section 2-8.
Special requirements over and above Graduate Division minimum
requirements
The IDP imposes no special
requirements.
Section 2-9.
Relationship of masters and doctoral programs
The M.S. and Ph.D. programs
are freestanding programs. Both
programs can admit and train students independent of each other. However, it is reasonable to permit students
to switch between programs. Thus,
M.S. students will be allowed to proceed to the Ph.D. program, subject to
approval by the IDP Director.
Similarly, Ph.D. students can receive the M.S. degree upon completion of
all its requirements.
Section 2-10.
Special preparation for careers in teaching
The IDP curriculum presents
numerous opportunities for graduate students to gain teaching experience. Students interested in teaching careers
will be strongly encouraged to gain experience as teaching assistants both in
undergraduate courses (e.g., Chemistry C160A) and graduate courses in
bioinformatics. All students in
the program must complete at least one quarter of teaching assistantship in
bioinformatics courses or in courses of participating departments.
Section 2-11.
Student sample program for each year
Ordinarily students will
complete the IDP curriculum in their first year.
Table 1.
Sample Student Program for the First Year
|
Fall |
Winter |
Spring |
|
Chem CM260A (Intro. to
Bioinformatics and Genomics) |
Chem C260B (Algorithms in
Bioinformatics and Systems Biology) |
Stats M254 (in future Chem
M260C?) (Statistical Methods in Computational Biology) |
|
elective |
Chem 260BL (Advanced
Bioinformatics Computational Laboratory) |
elective |
|
Chem 202 (Bioinformatics
Interdisciplinary Research Seminar) |
elective |
Chem 202 (Bioinformatics
Interdisciplinary. Research Seminar) |
|
Chem M252 (Seminar: Advanced Methods in Computational
Biology) |
Chem 202 (Bioinformatics
Interdiscip. Research Seminar) |
Chem M252 (Seminar: Advanced Methods in Computational
Biology) |
|
|
Chem M252 (Seminar: Adv. Methods in Computational
Biology) |
|
Ordinarily students would be
expected to complete the Oral Qualifying Exam and Bioinformatics Practicum
during the Summer quarter of their first year, and complete their Written
Qualifying Exam and Midstream Seminar by the end of their second year.
2-12.
Normative time from matriculation for degree (assuming student has no
deficiencies and is enrolled full-time)
2-12a. Normative lengths of time for
pre-candidacy and candidacy periods
We expect M.S. students to
complete the program within two years, and perhaps in as little as five
quarters. We expect Ph.D. students
to take two years to advance to candidacy, and three additional years to
complete the dissertation. This is
similar to the ACCESS program, whose average time to completion of the Ph.D.
degree is five years and one quarter.
2-12b. Other incentives to support expeditious
times-to-degree
The IDP Student Advising
Committee will conduct a yearly progress review of all students, based upon
coursework performance, research achievements, and faculty advisor
comments. Students who are not
progressing at the expected pace or evidently experiencing difficulties will be
contacted by the IDP Graduate Advisor to discuss the problem and recommend
solutions. Students who fail to make
adequate progress towards completion (as defined by the Graduate Handbook on
satisfactory academic progress) may be dismissed from the program. Students must complete the core
curriculum within the first two years.
Students normally must complete all course requirements by the end of
the second year (M.S.) or third year (Ph.D.). Ph.D. students should advance to candidacy by the end of
their third year. Failure to
advance to candidacy by the end of the fourth year will result in academic
probation.
Section 3. Projected Need
Section 3-1.
Student demand for the program
3-1a. Documentation
of demand for program with three to five years of enrollment and admissions
statistics from this or other institutions, or data on rate of student
inquiries
- UCLA established a bioinformatics curriculum in
1999. Since then we have
recommended to applicants interested in a Bioinformatics Ph.D. that they
apply to an existing UCLA graduate program (such as ACCESS) and then
complete the bioinformatics curriculum.
- In the two years that we recorded contacts (by
suggesting that interested applicants register via our online website), we
received inquiries from 315 students in 2000, and 961 students in
2001. Given that this number
of inquiries greatly exceeded our capacity to respond, we halted use of
the online inquiry system in 2002.
Table 2.
Enrollments of Bioinformatics Core Courses
|
Academic Year |
Course |
Final Enrollment |
|
1999-0 |
Chemistry 202 |
25 |
|
|
Chemistry 160/260 |
8 + 16 |
|
2000-1 |
Chemistry 202 |
13 |
|
|
Chemistry 160/260 |
21 + 20 |
|
2001-2 |
Chemistry 202 |
7 |
|
|
Chemistry 160/260 |
23 + 28 |
|
|
Statistics 254 |
34 |
|
2002-3 |
Chemistry 202 |
10 |
|
|
Chemistry 160/260 |
34 + 28 |
|
|
Statistics 254 |
30 |
|
2003-4 |
Chemistry 202 |
11 |
|
|
Chemistry 160/260 |
12 + 14 |
|
|
Statistics 254 |
19 |
|
2004-5 |
Chemistry 160/260 |
20 + 11 |
|
|
Statistics 254 |
22 |
|
2005-6 |
Chemistry 160/260 |
26 + 10 |
3-1b. Evidence
that this demand will be stable and long-lasting
There is
abundant evidence of a widespread and long-term commitment to bioinformatics
research.
- In the published literature: over the past few years several
bioinformatics scientific journals have been created. These include:
o
Briefings in Functional Genomics and
Proteomics
o
Computers in Biology and Medicine
o
Computer Methods and Programs in
Biomedicine
o
Journal of Biomedical Informatics
o
Journal of Computational Biology
- In NIH funding: the NIH has funded seven centers of computational
biology throughout the country.
One of these, the Center for Computational Biology, is located here
at UCLA.
- In faculty hiring: four of the bioinformatics core faculty have been hired
within the last year (Eskin, Pellegrini, Graber and Mallick), and one will
be coming to UCLA in Fall 2007 (Novembre).
- In establishment of major national laboratories
devoted to bioinformatics and genomics not only in the United States (the
National Center for Biotechnology Information, and the National Human
Genome Research Institute, both in Bethesda, MD), but also the European
Union (the European Bioinformatics Institute, with multiple sites).
- In growth of demand for bioinformatics databases
and queries. For example, the
National Center for Biotechnology Information is the top online
bioinformatics resource in the world. In 2000, its databases received 3,000,000
bioinformatics queries per day from users worldwide. In 2002, its databases processed
an average of more than 7,000,000 bioinformatics search requests per
day. This demonstrates the extremely
broad usage of bioinformatics tools not only by academic and industrial
researchers, but also by doctors, lawyers and even the public at large.
- In the creation of bioinformatics societies: e.g., The
International Society for Computational Biology, which organizes several international
bioinformatics meetings (ISMB and RECOMB).
3-1c. For
new programs that are extensions of existing disciplines, enrollment statistics
from related courses to demonstrate demand
Not
applicable.
3-1d. Statistics
or other documentation of need
See Section
3-2b below.
Section 3-2. Opportunities for placement of
graduates
3-2a. Placement
records of other UC programs in the field in recent years
This
data has not been available.
3-2b. Demonstration
of a strong market for program graduates by listing recent job listings,
employer surveys, assessments of future job growth
To demonstrate
the strong market demand for bioinformaticists, we list below the job openings
posted on the web site of the International Society for Computational Biology
for July through September of 2006.
There is a total of about 50 listings in both academia and industry.
|
2006-09-12 |
|
University
of Dundee, Scotland, UK |
|
|
2006-09-11 |
USA-SC-Columbia |
University
of South Carolina |
|
|
2006-09-11 |
USA-SC-Columbia |
University
of South Carolina |
|
|
2006-09-08 |
Singapore-Singapore |
Post-Doctoral
Research Fellow and Scientist: Computational Biologist/Bioinformatician |
Genome
Institute of Singapore |
|
2006-09-08 |
France-Paris |
Pasteur
Institute |
|
|
2006-09-08 |
France-Paris |
Pasteur
Institute |
|
|
2006-09-06 |
USA-ILLINOIS-Maywood |
Loyola
University Medical Center |
|
|
2006-09-05 |
USA-NJ-Newark |
UMDNJ-New
Jersey Medical School |
|
|
2006-09-01 |
France-Grenoble |
INRIA |
|
|
2006-08-30 |
USA-Arizona-Phoenix |
TGen |
|
|
2006-08-28 |
USA-MO-St.
Louis |
Post
doctorate researcher/research assistant in computational plant genome
analysis |
Donald
Danforth Plant Science Center |
|
2006-08-28 |
USA-MI-Ann
Arbor |
University
of Michigan |
|
|
2006-08-28 |
|
NYU School
of Medicine |
|
|
2006-08-25 |
45056-OH-Oxford |
Ph.D./M.S.
Assistantships in Plant Genomics and Bioinformatics |
Miami
University |
|
2006-08-23 |
USA-Delaware-Newark |
University
of Delaware |
|
|
2006-08-22 |
USA-CO-Boulder |
University
of Colorado at Boulder |
|
|
2006-08-22 |
South
Korea----Seoul |
Research
Center for Women’s Diseases, Sookmyung Univ. |
|
|
2006-08-20 |
|
Postdoc
positions in microbial pathway/network modeling and simulation |
University
of Georgia |
|
2006-08-17 |
USA-CA-La
Jolla |
UCSD |
|
|
2006-08-14 |
USA-Iowa-Ames |
Iowa State
University |
|
|
2006-08-14 |
Iowa-Ames |
Iowa State
University |
|
|
2006-08-14 |
Iowa-Ames |
Iowa State
University |
|
|
2006-08-09 |
Italy-Tuscany-Siena |
Siena
Biotech S.p.A. |
|
|
2006-08-09 |
Italy-Tuscany-Siena |
Siena
Biotech S.p.A. |
|
|
2006-08-08 |
US-Massachusetts-Boston |
Harvard
Medical School / Children’s Hospital |
|
|
2006-08-04 |
France-Lyon |
French
Institute of Computer Science (INRIA) |
|
|
2006-08-01 |
USA-California-Los
Angeles |
Loyola
Marymount University |
|
|
2006-07-30 |
USA-California-Los
Angeles |
University
of Southern California |
|
|
2006-07-28 |
TX-Houston |
University
of Houston |
|
|
2006-07-27 |
USA-GA-Athens |
University
of Georgia |
|
|
2006-07-27 |
USA-Georgia-Atlanta |
Centers for
Disease Control and Prevention |
|
|
2006-07-26 |
USA-NC-Chapel
Hill |
University
of North Carolina at Chapel Hill |
|
|
2006-07-26 |
USA-MA-Boston |
Boston
College |
|
|
2006-07-26 |
Canada-ON-Toronto |
Postdoctoral
positions in computational biology/bioinformatics of cellular networks |
University
of Toronto |
|
2006-07-24 |
USA-Michigan-East
Lansing |
Faculty
Positions in Plant Proteomics and Quantitative Biology |
Michigan
State University |
|
2006-07-24 |
United
Kingdom-Berkshire-Bracknell |
Syngenta
Crop Protection AG |
|
|
2006-07-23 |
USA-NY-New
York |
Weill
Medical College of Cornell University |
|
|
2006-07-20 |
USA-New
York-New York |
Memorial
Sloan-Kettering Cancer Center |
|
|
2006-07-18 |
USA-OH-Oxford |
Miami
University |
|
|
2006-07-17 |
USA-AR-Jefferson |
Z-Tech
Corporation |
|
|
2006-07-14 |
USA-Wisconsin-Madison |
Center for
Eukaryotic Structural Genomics--Univ. of Wisconsin |
|
|
2006-07-13 |
USA-CA-Palo
Alto |
Stanford
University School of Medicine |
|
|
2006-07-10 |
USA-Maryland-Baltimore |
Postdoctoral
fellowship in computational biology of angiogenesis |
Johns
Hopkins University |
|
2006-07-07 |
USA-GA-Athens |
University
of Georgia |
|
|
2006-07-07 |
Canada-ON-Ottawa |
Tenure-track
faculty appointment in the area of biomedical engineering or bioinformatics |
University
of Ottawa |
|
2006-07-05 |
Canada-BC-Vancouver |
University
of British Columbia (Vancouver) |
|
|
2006-07-03 |
USA-CA-Irvine |
University
of California, Irvine |
|
|
2006-07-01 |
France-Bordeaux |
LaBRI,
University of Bordeaux |
|
|
2006-06-28 |
USA-Kansas
City |
Stowers
Institute for Medical Research |
|
|
2006-06-26 |
USA-TX-Houston |
The
University of Texas M. D. Anderson Cancer Center |
|
|
2006-06-22 |
Japan-Yamagata-Tsuruoka |
Keio
University / E-Cell Project |
|
|
2006-06-19 |
USA-CA-Santa
Barbara |
University
of California at Santa Barbara |
|
|
2006-06-16 |
|
Johns Hopkins |
|
|
2006-06-16 |
USA-MD-Rockville |
The
Institute for Genomic Research |
|
|
2006-06-16 |
USA-MD-Rockville |
The
Institute for Genomic Research |
|
|
|
|
|
|
Section
3-3. The importance of the new
program to the discipline
UCLA will help
prepare the next generation of bioinformatics scientists, emphasizing core
strengths that are needed by the field.
First, UCLA provides strongly integrated training that brings together
perspectives (genomics, proteomics, computer science, mathematics and
statistics) that are often distant from each other in the existing
bioinformatics community. The fact
that UCLA brings together many bioinformatics faculty in each of these four
areas, joining together the full power of world-class schools in medicine,
engineering, and life and math/physical sciences, is a major strength
replicated in very few universities worldwide. Second, UCLA has translated this strong integration and
collaboration from its research into its graduate training. Our students receive explicit training
in the skills of interdisciplinary collaboration, and will be able to cross
academic boundaries and collaborate with bioinformatics researchers from other
backgrounds.
These skills
are important for the field of bioinformatics because it is new, requires
interdisciplinary collaboration, and has a serious shortage of scientists who
are able to work successfully across the boundaries of fields as distant as
molecular biology and computer science.
Bioinformatics is a brand-new field; most of the fundamental science
needed to deliver on its promises (e.g., the ability to understand how the
human genome encodes the capabilities of the human mind) has yet to be
invented. Until researchers from
many different backgrounds are able to work together in both a collaborative
and creative way, these incredible challenges will remain unsolved. Currently, there is a shortage of
bioinformatics scientists who are able to bridge such gaps in the way that is
needed. This need has been pointed
out by study after study at both institutional and national levels. We believe that UCLA can make a
valuable contribution to this need.
Section 3-4. Ways in which the program will meet the
needs of society
As technology
advances, experimental techniques used in biomedical labs will continue to
generate more and more data. While
only a decade ago a typical experiment generated only a handful of data points,
current experimental techniques generate thousands. Furthermore, there are rapidly growing repositories of this
data throughout the web. This
revolution is changing the very nature of biomedical research. It is becoming increasingly difficult
for young scientists to make important contributions in their field unless they
are adept at analyzing and extracting information from this data.
Bioinformatics
training is therefore a critical component of our scientific training. Without a strong foundation in this
field, our society will no longer be able to compete and make contributions in
biomedical research. It is clear
not only that the IDP in bioinformatics proposed here will contribute to
enhancing the scientific base of a few graduate students at UCLA, but that this
training will have far reaching implications for the competitiveness of biomedical
research in the US over the next few decades.
Section 3-5. Relationship of the program to research
and/or professional interests of the faculty
3-5a. Core
faculty
- James Bowie: Chemistry
& Biochemistry – computational methods to predict
membrane protein structure; analysis of primary sequence determinants of
membrane protein structure.
- Joseph DiStefano: Computer
Science –
systems biology.
- David Eisenberg: Chemistry
& Biochemistry – protein interaction networks;
structural genomics; prediction of protein folding by 3D profile methods;
comparison and analysis of protein structures, and protein
sequence-structure relationships.
- Eleazar
Eskin: Computer
Science –
computational analysis of genetics and genomics data.
- Tom Graeber: Molecular & Medical
Pharmacology – understanding cancer signaling from a
systems viewpoint.
- Steve Horvath: Biostatistics – allelic association and DNA microarray
data analysis. Identification and characterization of disease genes.
- James Lake: Molecular,
Cell & Developmental Biology – bioinformatics analyses of genome
evolution, focusing on the evolution and origins of eukaryotes and
prokaryotes.
- Ken Lange: Biomathematics – computational genetics, population
genetics, gene mapping, radiation hybrids, pedigree analysis, biomedical
imaging, computational statistics, applied stochastic processes.
- Christopher Lee: Chemistry
& Biochemistry – evidence-confidence computation for
genome-wide wide analyses of raw experimental data. High-throughput discovery of
single- nucleotide polymorphisms, alternative splicing, and gene
structure. Proteome
annotation and databases.
- Ker-Chau
Li: Statistics – high dimensional data analysis.
- James Liao: Chemical
Engineering –
intracellular regulation and metabolism at the systems level.
- Parag Mallick:
director of clinical proteomics at Cedars-Sinai Medical Center and
Chemistry and Biochemistry – bioinformatics with a focus on mass
spectrometry.
- John Novembre: Ecology
and Evolutionary Biology (Fall 2007) -- genetic variation within species
and genomic divergence between species
- Stott
Parker:
Computer Science – databases and data-mining. Novel database architectures for
genomics.
- Matteo Pellegrini: Molecular, Cell and Developmental Biology – development of computational approaches
to model and interpret genomic data with a focus on the reconstruction of
molecular networks.
- Chiara Sabatti: Statistics – statistical genetics, Markov Chain
Monte Carlo, Bayesian statistics.
- Janet Sinsheimer: Human
Genetics –
statistical modeling of evolution and genetics.
- Marc Suchard: Biomathematics – statistical methodology; genetics;
evolution.
- Todd Yeates: Chemistry
& Biochemistry – structure proteomics - working to discover
and determine new protein folds.
Protein evolution - investigating unusual mechanism of protein
evolution. Functional
genomics – working on methods to infer function from sequence.
- Qing
Zhou: Statistics – development of statistical methods in
cis-regulatory analysis, including motif and cis-regulatory module
discovery, gene expression analysis, ChIP-chip data analysis, and
comparative genomics.
3-5b. Associate
faculty
- Guillaume Chanfreau: Chemistry
& Biochemistry – RNA structure and processing.
- Ivo
Dinov: Statistics – brain image analysis.
- Fred Fox: Microbiology,
Immunology and Molecular Genetics – bioinformatics in industry; industrial
contacts.
- Siavash Kurdistani: Biological
Chemistry – genomic
studies of chromatin biology.
- Ralf Landgraf: Biological
Chemistry –
structure-function relationships of macromolecular interactions.
- Joseph Loo: Chemistry
and Biochemistry –
mass spectrometry.
- Stanley Nelson: Human
Genetics –
DNA microarrays.
Identification and characterization of disease genes
- Peipei Ping: Physiology,
Medicine, and Cardiology – functional proteomic analysis of the normal,
ischemic and protected myocardium.
- Stefano
Soatto: Computer
Science –
image processing and shape analysis.
- Charles Taylor: Ecology
and Evolutionary Biology – population genetics and adaptation;
population structure of malaria vectors; adaptation in sensor arrays,
especially for bioacoustics.
- Arthur Toga: Neurology – brain imaging.
- Yingnian
Wu: Statistics
– image
modeling, statistical computing.
Section 3-6. Differentiation of the proposed program
from existing UC and California independent university programs, and from
similar programs proposed by other UC campuses
The program we
propose here is focused on providing skills to perform innovative
bioinformatics research. This will
involve both basic and applied research.
Some of the other programs in California universities focus more on medical
data and therefore more on applied research. Other programs combine informatics with experimental
techniques used for genomic research (e.g., Riverside). Other programs do not provide a Ph.D.,
and therefore mainly provide an addition to existing graduate programs (e.g.,
UC Berkeley).
The key points
that differentiate our program from others are therefore:
- A large list of hardcore bioinformatics faculty
unifying the approaches of four different schools: Medicine (Human
Genetics; Biomathematics; Biostatistics; MMP); Engineering (Computer
Science; Chemical Engineering); Life Sciences (MCDB; EEB); Physical
Sciences (Statistics; Chemistry).
- A rigorous, integrated core curriculum teaching
the fundamental principles for inventing new kinds of bioinformatics far
into the future.
- A Bioinformatics Ph.D., and a Masters program,
designed for students with a bioinformatics / computational biology
undergraduate degree, or equivalent high level of preparation for the
field.
We believe
that these are the critical components that will make this program attractive
to undergraduate students focused on pursuing a research career in
bioinformatics and distinct from other programs offered by California
universities.
Section 4. Staff
Section 4-1. Program Faculty
There are 20 core faculty
involved in teaching the required and elective courses of the proposed
IDP. The core faculty are listed
in Section 3-5a, and associate faculty are listed in Section 3-5b. Faculty
hiring plans are described in Section 1-3a.
4-1a. List of program faculty, their ranks,
their highest degree and other professional qualifications, and a citation of
no more than five recent publications (abbreviated curricula vitae);
Data concerning faculty limited to that information pertinent to the
committee’s evaluation of faculty qualifications
See Section 3-5 above. Brief curriculum vitae of core and
associate faculty are in Appendix C.
4-1b. Comments from chairs of departments with
graduate programs and/or faculty closely related to or affected by the proposed
program
See Appendix D.
4-1c. For participating faculty members outside
of the sponsoring department, copies of letters indicating their interest in
the program (critical for interdisciplinary programs)
See Appendix E.
Section 4-2. Organizational Structure
The Chair of the IDP will be recommended to the Dean
of the Graduate Division by the Dean of Life Sciences, as specified by the
regulations of the Academic Senate that govern the appointment of IDP
chairs. The Executive Graduate
Program Committee for the IDP will be constituted of no less than four members,
each representing one of the participating Schools or Divisions –
Medicine, Engineering, Life Sciences and Physical Science. Executive Committee members will
normally serving a three year term.
Members of the committee will be recommended to the Dean of the Graduate
Division by the Chair of the IDP and the Dean representing the home Division of
School of the Executive Committee member.
Additional members may be recommended to the Dean of the Graduate
Division by agreement of the Chair of the IDP and the IDP Executive
Committee. Members will be
selected to provide broad representation of the areas of specialization. The duties of the Graduate Program
Committee will be to determine policy for the administration of the IDP, select
students for admission to the IDP, review and approve course selections for
meeting each student's requirement for coursework, and approve selection of the
student's advisory committee. The
IDP will be evaluated and advised by a UCLA Faculty Advisory Committee with
representatives from academic units outside Life Sciences, an External Advisory
Board, and an Industrial Advisory Board.
The proposed organizational structure of the IDP is illustrated below:
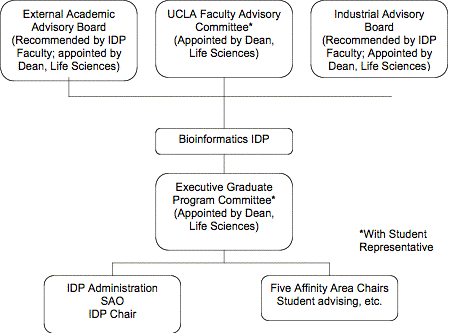
Section 4.3 Sources of available fellowship and
assistantship support
Doctoral students will be supported through intra and
extramural resources including teaching assistant appointments through Life
Science, Physical Science or Engineering, unrestricted fellowship funds
provided through formulaic allocations to the IDP from the Graduate Division,
institutional NRSA training grants including but not restricted to Cell and
Molecular Biology, Genetics, Biotechnology, and Genomic Analysis, as well as
through GSR appointments from research grants obtained by their mentors. See Section 7 for details.
Section 5. Courses
Section 5-1. Present and proposed courses, with
planned instructors, and supporting courses offered in related fields
Table 3. Present and Proposed Courses for the Program
|
Course Title |
Instructors |
Status |
Major Area |
Schedule |
|
Core Curriculum |
|
|
|
|
|
Introduction to
Bioinformatics & Genomics (Chem CM260A) |
Lee (Chem) |
Existing |
Core |
Fall, annually |
|
Advanced Algorithms in
Bioinformatics (Chem C260B) |
Mallick (Chem) |
Existing (taught W07) |
Core |
Winter, annually |
|
Bioinformatics Algorithms
Laboratory (Chem 260BL) |
Eskin (CS), Mallick (Chem) |
Pending (Winter 2008) |
Core |
Winter annually |
|
Statistical Methods in
Computational Biology (Stat 254/Chem 260C) |
Sabbati (Hum Gen /
Statistics) |
Existing |
Core |
Spring, annually |
|
Bioinformatics
Interdisciplinary Research Laboratory (Chem 202) |
Lee (Chem) Pellegrini (MCDB), core
faculty |
Existing |
Core |
Fall, Winter, Spring,
annually |
|
Additional Required
Courses |
|
|
|
|
|
Computational Biology
Research Seminar (Chem M252) |
Eisenberg (Biol Chem) |
Existing |
Required |
Fall, Winter, Spring,
annually |
|
Laboratory Rotation (596) |
Core faculty |
Existing |
Required for Ph.D. |
Every quarter |
Section
5-2. Elective Bioinformatics
Courses
Full course catalog
descriptions of all required and elective courses are included in Appendix F.
Ecology and Evolutionary
Biology M231. Molecular Evolution. (Same
as Earth and Space Sciences M217.) Lecture, two hours; discussion, two hours.
Series of advanced topics in molecular evolution, with special emphasis on
molecular phylogenetics. Topics may include nature of the genome, neutral
evolution, molecular clocks, concerted evolution, molecular systematics,
statistical tests, and phylogenetic algorithms.
Biomath M203. Stochastic Models in Biology. (Same as
Human Genetics M203.) Lecture, four hours. Requisite: Mathematics 170A or
equivalent experience in probability. Mathematical description of biological
relationships, with particular attention to areas where conditions for
deterministic models are inadequate. Examples of stochastic models from
genetics, physiology, ecology, and a variety of other biological and medical
disciplines.
Biomath M207A. Theoretical Genetic Modeling (Formerly
numbered 207.) (Same as Biostatistics M237A.) Lecture, three hours; discussion,
one hour. Requisites: Mathematics 115A, 131A, Statistics 100B Mathematical models in statistical
genetics. Topics include population genetics, genetic epidemiology, gene
mapping, design of genetics experiments, DNA sequence analysis, and molecular
phylogeny.
Biomath 210. Optimization Methods in Biology. Lecture, four hours. Preparation: undergraduate
mathematical analysis and linear algebra; familiarity with programming language
such as FORTRAN or C. Modern computational biology relies heavily on
finite-dimensional optimization. Survey of theory and numerical methods for
discrete and continuous optimization, with applications from genetics, medical
imaging, pharmacokinetics, and statistics.
Biomath M211. Mathematical
and Statistical Phylogenetics. (Same as Human Genetics M211.) Lecture, three
hours; laboratory, one hour. Requisites: Biostatistics 110A, 110B, Mathematics
170A. Theoretical models in molecular evolution, with focus on phylogenetic
techniques. Topics include evolutionary tree reconstruction methods, studies of
viral evolution, phylogeography, and coalescent approaches. Examples from
evolutionary biology and medicine. Laboratory for hands-on computer analysis of
sequence data.
Biostatistics M272.
Theoretical Genetic Modeling. (Formerly numbered M237A.) (Same as
Biomathematics M207A and Human Genetics M207A.) Lecture, three hours;
discussion, one hour. Requisites: Mathematics 115A, 131A, Statistics 100B.
Mathematical models in statistical genetics. Topics include population
genetics, genetic epidemiology, gene mapping, design of genetics experiments,
DNA sequence analysis, and molecular phylogeny.
Biostatistics M278. Statistical Analysis of DNA Microarray
Data. (Formerly numbered 278.) (Same as Human Genetics M278.) Lecture, three
hours. Requisite: course 200C. Instruction in use of statistical tools used to
analyze microarray data. Structure corresponds to analytical protocol an
investigator might follow when working with microarray data.
Chemistry 298. Seminar: Computational Methods in
Bioinformatics. To be proposed.
Discussion, one hour. Detailed analysis of new methods, including
algorithms and representation of bioinformatics data, and research problems
ranging from genome-wide sequence analysis, to fold recognition, to energy
calculation and protein design.
Computer Science 249. Current
Topics in Data Structures. Lecture, four hours; outside study, eight hours.
Review of current literature in an area of data structures in which instructor
has developed special proficiency as a consequence of research interests.
Students report on selected topics.
Human Genetics 244. Genomic Technology. Lecture, three
hours; discussion, one hour. Requisite: Life Sciences 4. Survey of key
technologies that have led to successful application of genomics to biology,
with focus on theory behind specific genome-wide technologies and their current
applications. Concurrently scheduled with course C144.
MCDB 292. Seminar: Molecular Evolution.
Discussion, three hours. Detailed analysis of current understanding of evolution
of molecular sequences and structures.
Pathology 255. Mapping and Mining Human Genomes.
Lecture, 90 minutes; discussion, 90 minutes. Basic molecular genetic and
cytogenetic techniques of gene mapping. Selected regions of human genomic map
scrutinized in detail, particularly gene families and clusters of genes that
have remained linked from mouse to human. Discussion of localizations of
disease genes.
Statistics 165: Statistical Methods and Data
Mining. Lecture, three hours.
Requisite: course 100A. Introduction and overview of up-to-date statistical
methods in microarray analysis designed for students in biostatistics,
statistics, and human genetics who are interested in technology and statistical
analysis of microarray experiments. Useful for biology students with basic
statistical training who are interested in understanding logic underlying many
statistical methods.
Neuroinformatics
Neuroscience/Physiological
Science CM272. Neuroimaging and Brain Mapping. (Same as Physiological Science
M272 and Psychology M213.) Lecture, three hours. Requisites: courses M201,
M202. Theory, methods, applications, assumptions, and limitations of
neuroimaging. Techniques, biological questions, and results. Brain structure,
brain function, and their relationship discussed with regard to imaging.
Statistics 233 - Statistical
Methods for Biomedical Imaging.
Lecture, three hours. Requisite: course 100A. Brief review of common
general statistical techniques. Advanced statistical methods for analysis of medical
imaging, integration, visualization, interrogation, and interpretation of
imaging and nonimaging metadata.
(http://www.stat.ucla.edu/%7Edinov/courses_students.dir/04/Spring/Stat233.dir/STAT233.html)
Biomedical Physics 208A.
Medical Physics Laboratory: Medical Imaging. Discussion, two hours; laboratory,
four hours. Requisite: course 205.
Hands-on experience performing acceptance testing and quality control
checks of imaging equipment such as fluoroscopy, digital subtraction
angiography, mammography, ultrasound, magnetic resonance imaging, computed
tomography, and computed radiography.
Biomedical Physics 210.
Principles of Medical Imaging. Lecture, three hours; discussion, one hour.
Requisite: course 209. Study of image representation, computational structures
for imaging, linear systems theory, image enhancement and restoration, image
compression, segmentation, and morphology. Special topics include visualization
techniques, three-dimensional modeling, computer graphics, and neural net
applications. Laboratory projects apply concepts developed in class.
Biomedical Physics 214.
Medical Image Processing Systems. Lecture, three hours; discussion, one hour. Requisites:
courses 209, 210. Advanced image processing and image analysis techniques
applied to medical images. Discussion of approaches to computer-aided diagnosis
and image quantitation, as well as application of pattern classification
techniques (neural networks and discriminant analysis). Examination of problems
from several imaging modalities (CT, MR, CR, and mammography).
Biomedical Physics M266.
Advanced Magnetic Resonance Imaging. (Same as Neuroscience M267 and Psychiatry
M266.) Lecture, four hours. Starting with basic principles, presentation of
physical basis of magnetic resonance imaging (MRI), with emphasis on developing
advanced applications in biomedical imaging, including both structural and
functional studies. Instruction more intuitive than mathematical.
Biomedical Physics M285.
Functional Neuroimaging: Techniques and Applications. (Same as Psychiatry
M285.) In-depth examination of activation imaging, including PET and MRI
methods, data acquisition and analysis, experimental design, and results
obtained thus far in human systems. Strong focus on understanding technologies,
how to design activation imaging paradigms, and how to interpret results.
Laboratory visits and design and implementation of a functional MRI experiment.
Biomedical Engineering M217.
Biomedical Imaging. (Same as Electrical Engineering M217.) Lecture, three
hours; laboratory, two hours; outside study, seven hours. Requisite: Electrical
Engineering 114D or 211A. Mathematical principles of medical imaging
modalities: X-ray, computed tomography, positron emission tomography, single
photon emission computed tomography, magnetic resonance imaging. Topics include
basic principles of each imaging system, image reconstruction algorithms,
system configurations and their effects on reconstruction algorithms,
specialized imaging techniques for specific applications such as flow imaging.
The following supporting courses are
offered in related fields: Statistics 100A (or equivalent
preparation) is required as a prerequisite for Chemistry CM260A. CS 31 (or equivalent programming
skills) is required for Chemistry C260BL (Bioinformatics Algorithms
Laboratory). The Program in
Computing offers a range of courses (PIC 10ABC, PIC 20AB, PIC 60, PIC 110) that
are also very useful for bioinformatics students. The many departments of the Medical School and Life Sciences
offer a wide range of coursework on biology that is highly relevant to
bioinformatics students.
Section 5-3. Program degree requirements
See Appendix G.
Section 6. Resource Requirements and Enrollment Plans
Section 6-1.
Methods for funding
Bioinformatics research has been an integral part of
the Life Science, Physical Science, Medical and Engineering departments at UCLA
for several years. The development
of this degree program utilized resources already available to faculty and
research programs without significant additional costs.
6-1a. FTE faculty
The core
faculty, consisting of 20 existing FTE, will teach, administer and support the
activities of the proposed IDP. As
documented in Sections 4 and 5, the existing faculty is more than adequate to
provide this graduate program at its project enrollment levels. UCLA is indeed fortunate to have a
group of bioinformatics faculty already assembled and working together. As early as 1999, a strategic planning
group for the Schools of Medicine, Life Sciences, and Physical Sciences, under
the direction of Deans Levey, Eiserling and Peccei, recommended that “the
highest research priority area that emerged from this process is
bioinformatics. We have interacted
considerably on these issues and we feel strongly also that this area should be
UCLA’s highest priority.” The
strength of the current bioinformatics core faculty demonstrates UCLA’s success
in achieving this goal. A variety
of departments have made recent hires of new faculty working in the field of
bioinformatics; for example, in the last two years: Ralf Landgraf (Biological Chemistry), Matteo Pellegrini
(MCDB), Thomas Graeber (Molecular and Medical Pharmacology), Parag Mallick
(Chemistry and Biochemistry), Eleazar Eskin (Computer Science), and John
Novembre (Ecology and Evolutionary Biology). Long-term campus plans have called for hiring several
additional faculty members in bioinformatics over the next five years. Startup costs for each new faculty hire
are likely to average $500,000, with annual salaries plus benefits of $100,000.
6-1b. Staff
FTE
The program
will be administered by existing student affairs staff (Student Affairs Officer
II) in the Department of Molecular, Cell and Developmental Biology and the
umbrella unit, Life Sciences South Administration (MSP I, Functional Manager)
in addition to current duties. It
is anticipated that less that 50% FTE will be required to provide
administrative support for student counseling, admissions processes and IDP
administration.
6-1c. Library
acquisitions
The Library’s
current holdings are sufficient to support the proposed
program. Bioinformatics, biology and computer science journals are
largely available on-line via the Worldwide Web. In addition to major initiatives such as PubMed Central that
are providing universal free access to a large number of journals in the field,
the UCLA Library has negotiated extensive online access agreements that cover
virtually all of the journals in the bioinformatics field. The current situation regarding journal
access is completely satisfactory for the needs of the proposed IDP.
Regarding library holdings,
it would be helpful to select one library on campus to act as a center for
bioinformatics holdings. However,
this is by no means a critical issue for creation of the IDP.
No
retrospective materials (monographic and serial fills) are required for
graduate level research. Bioinformatics research is distributed
via two mechanisms: publication in peer-reviewed journals; and via the Internet
(e.g., making software available on a website for download by other
researchers, or creating an online database which researchers can access over
the Internet).
The Library
currently subscribes to the most important serial publications (print or
electronic) in the field. Current subscriptions and journal
online access agreements are entirely satisfactory for the needs of the IDP.
6-1d. Computing
costs
Computing costs constitute a major
component of research expenditures, and have been paid from several sources in
the past: 1) faculty research grants; 2) private foundation grants (e.g.,
grants from the Keck Foundation and Sun Microsystems); 3) multi-investigator
“center” or training grants. The
most recent equipment award was a $200,000 supplement in 2002 from UCLA’s
NSF-IGERT, for shared computing equipment for IGERT Bioinformatics
students. Grant applications for
replacing or upgrading computer facilities will be necessary in the
future. The projected number of
new student enrollments under this program does not pose a serious additional
burden on existing computing resources.
6-1e. Equipment—inventory
of current equipment and future needs
UCLA Bioinformatics has many
years of experience building and running powerful clusters of low-cost LINUX PC
nodes. Currently, the computing
resources of the IDP faculty include over 1000 CPUs and approximately 50
terabytes of RAID disk storage.
These needs are driven primarily by the faculty’s research requirements,
not by IDP training requirements.
As noted above in Section 6-1d, we already have sufficient computer
power specifically for the projected IDP enrollment.
As one example, UCLA’s NSF
IGERT training grant provided funds for establishment of a 150 CPU computer
cluster specifically for bioinformatics graduate students. The Dean of Physical Sciences supplied
$30,000 for installation of sufficient air conditioning for the heat load of this
cluster. Space for the IGERT
cluster (in Young Hall 2069) was provided by the Department of Chemistry and
Biochemistry.
6-1f. Space
and other capital facilities—inventory of current facilities and future
requirements
No new space or capital
facilities will be required for this program beyond that already used by
program faculty.
6-1g. Other
operating costs (technical and administrative staff, supplies and expenses, lab
maintenance and other facilities) and description of current staffing levels
and future requirements
Funds to support the periodic
production of recruitment materials and development of a speakers program will
be coordinated through the Dean of Life Sciences with appropriate level
contributions solicited from Medicine, Engineering and Physical Sciences.
Section 6-2. Projected doctoral enrollments for the
first five years
Table 4.
Projected Doctoral Enrollments
|
Year |
New |
Continuing |
Total |
|
1 |
3 |
0 |
3 |
|
2 |
5 |
3 |
8 |
|
3 |
5 |
8 |
13 |
|
4 |
5 |
13 |
18 |
|
5 |
5 |
18 |
23 |
Section 7. Graduate Student Support
Section 7-1.
Strategy for meeting support needs
UCLA has
already secured many sources of funding needed for supporting both the
projected graduate enrollments.
Historically, UCLA has been very successful in competing for newly
created research and training grants for bioinformatics:
- In 2000, UCLA was one of two universities
nationwide to be awarded one of the first large-scale training grants in
bioinformatics from the National Science Foundation, the Integrated
Graduate Education and Research Training (IGERT). This $3 million, five-year grant
funded up to 15 Ph.D. students per year.
- In 2001, UCLA was awarded a large-scale center
grant in computational biology from a new program established by the
National Institutes of Health, the National Programs of Excellence in
Biomedical Computing.
- In 2001, UCLA was awarded a training grant to
establish a M.S. degree in Bioinformatics by the Sloan Foundation. This $100,000, three-year grant
supported up to five M.S. students per year.
- In 2002, UCLA was awarded a large training grant
in statistical genetics and genomics by the National Institutes of Health
(Genomic Analysis and Interpretation Training Grant, Ken Lange, PI). This five-year grant supports up
to 12 Ph.D. students per year.
- In 2004, UCLA was awarded a five-year, $20
million grant to establish the Center for Computational Biology, one of
the first four National Centers for Biomedical Computing, as part of NIH’s
Roadmap Initiative in Bioinformatics and Computational Biology. This grant supports both research
and training.
- In 2004, UCLA received a five-year, $5 million
grant for the UCLA-DOE Laboratory, which officially changed its name to
the Institute for Genomics and Proteomics to emphasize genome and proteome
analysis using bioinformatics.
- In 2005, UCLA was awarded a Biotechnology
training grant by NIH (Hal Monbouquette, PI).
- In addition to these multi-investigator training
grants, the faculty of the IDP pay for graduate student support directly
from individual research grants awarded by public agencies and private
foundations.
The Bioinformatics
IDP’s strategy for funding Ph.D. student support has the following elements:
- Admitted Ph.D. students will be offered
guaranteed funding for at least one year from the above funding sources.
- After the first year of rotations, each Ph.D.
student joins the laboratory of an IDP faculty member. From this point on, students
usually are supported by a competitive training grant or from the faculty
advisor’s research grants.
- Bioinformatics graduate training is a high
priority area for many agencies that offer individual graduate fellowship
awards. We will strongly
encourage our top students to apply for outside graduate fellowships, such
as the NSF Graduate Research Fellowship; DOE Computational Science
Graduate Fellowship; HHMI Predoctoral Fellowship, etc.
- UCLA Graduate Division provides funding for
graduate support. The
Graduate Division provides funds to programs according to the number of
enrolled students. Based on
current allocation levels, we anticipate an annual allocation of
approximately $75,000 when the program reaches steady state.
- Regarding available teaching assistants and
resources for TAs through enrollment growth, reallocation, or combination,
and the effect of any reallocations on support in existing graduate
programs: as described above, TA support is not needed as a major element of
graduate student support for the proposed IDP. Some IDP students will work as Teaching Assistants in
bioinformatics courses and in some affiliated undergraduate courses
(Chemistry 160). Currently,
these total about four to five TAships per year, but are likely to
increase (up to 15 TAships per year) once the IDP begins operation. This will come from increased TA
allocations due to increased bioinformatics course enrollments, from
development of new undergraduate courses in bioinformatics and
computational biology, and from existing course offerings in associated
departments. This will have a
minimal impact on support in existing graduate programs.
Section 7-2.
Current availability of faculty grants to support graduate
students and funding trends in agencies expected to provide future research or
training grants
The National
Institutes of Health (NIH) is the major source of funding for biomedical
research in the U.S. NIH has
prioritized Bioinformatics as one of the most important areas for new grants
for research and education; indeed it was one of the first four fields targeted
under the NIH Roadmap Initiative.
To carry out this mandate, NIH established the Biomedical Information
Science and Technology Initiative (BISTI). Since its inception in 2000, BISTI and other NIH institutes
have launched over 35 distinct funding programs in bioinformatics (see www.bisti.nih.gov). The pace of new funding program
announcements has accelerated in recent years: 1 in 1998; 2 in 2000; 4 in 2001;
8 in 2002; and 15 in 2003. These
bioinformatics funding programs now involve nearly all of the individual
Institutes of NIH, demonstrating that the need for bioinformatics is perceived
as essential for all areas of biomedical research from basic science, to
clinical studies of the genetics of disease, to brain science and mental
health.
For example,
just one BISTI program, the National Centers for Biomedical Computing (NCBC),
calls for the creation of up to 20 centers totaling approximately $400 million
in the first five years of funding.
UCLA was awarded one of the very first center grants (a five-year grant
totaling $20 million). So far, in
the first two years of the National Centers program, seven centers have already
been established. Following the
model of the NCBC program, BISTI has launched a number of new center programs
targeting bioinformatics: Centers
of Excellence in Genomic Science; Centers of Excellence in Complex Biomedical
Systems Research; Core Centers for Advanced Neuroinformatics Research; National
Centers for Systems Biology.
Other federal
agencies have also identified bioinformatics area as a key area of research,
and have established grant programs to support bioinformatics research and
training.
- NSF has established a large program of grant
support for bioinformatics research and bioinformatics resources. For example, through its
Biological Databases and Informatics program and associated grants, NSF
has made 97 grants totaling $64 million since its inception in 2000. Many other NSF grant programs
include a large bioinformatics component. For example, NSF established the Integrated Graduate
Education and Research Traineeship program (IGERT) to foster
interdisciplinary graduate training, and bioinformatics was a significant
focus from the beginning. In
fact, UCLA was awarded one of the first three IGERT grants, a five-year,
approximately $3 million grant for Ph.D. training in bioinformatics.
- DOE
- Howard Hughes Medical Institute
- Many private foundations offer research and
training grants for bioinformatics.
For example, the Pharmaceutical Research and Manufacturers of
America Foundation (PhRMA) has targeted bioinformatics as one of six areas
it is funding through its grants program.
Section 7-3.
Other extramural resources likely to provide graduate student support,
or internal fellowships and other institutional support made available to the
program
Bioinformatics
students already receive support from a number of externally funded training
grants at UCLA, and students enrolled in the IDP will be eligible for the
following grants: the Genomic Analysis and Interpretation Training Grant (K.
Lange, PI); the Training Program in Genetic Mechanisms (R. Simons, PI); and
Biotechnology Training in Biomedical Sciences and Engineering Program (H.
Monbouquette, PI).
Bioinformatics
graduate training is a high priority area for many agencies that offer
individual graduate fellowship awards (e.g., Howard Hughes Medical Institute,
National Science Foundation, Department of Energy). We will strongly encourage our top students to apply for
outside graduate fellowships.
Section 7-4. Campus fund-raising initiatives that
will contribute to support of graduate students
The Bioinformatics IDP
faculty have participated in a number of successful campus fund-raising
initiatives, including:
- Keck Center for Proteomics: in 2000, the Keck Foundation
awarded UCLA a $5 million grant to establish a proteomics laboratory,
Bioinformatics User Facility and genomics core facilities.
- Howard Hughes Medical Institute: in 2000, HHMI awarded the UCLA
School of Medicine a $2.1 million Research Resource Grant for
bioinformatics and other research core facilities.
- Sun Microsystems: in 2000, Sun contributed approximately $1 million in
enterprise computing equipment to the UCLA Bioinformatics program.
Currently, the IDP faculty
and Deans of Medicine, Life Sciences, and Physical Sciences are developing a
fund-raising initiative in genomics, which may include a component for
bioinformatics graduate student support.
In addition, the recently created EUREKA endowment for graduate student
support has raised approximately $3 million of a planned $40 million endowment
fund. Bioinformatics IDP students
will be eligible for these funds.
Section 7-5.
Graduate student support table listing maximum number of students
projected and sources of support for the first six years of the program
Table 5.
Graduate Student Support
|
Year |
Students |
Source of Support |
|
1 |
3 |
Graduate Division NRSA Training Grants |
|
2 |
8 |
Graduate Division NRSA Training Grants Teaching Assistantships Extramural Research Grants |
|
3 |
13 |
Graduate Division NRSA Training Grants Teaching Assistantships Extramural Research Grants |
|
4 |
18 |
Graduate Division NRSA Training Grants Teaching Assistantships Extramural Research Grants |
|
5 |
23 |
Graduate Division NRSA Training Grants Teaching Assistantships Extramural Research Grants |
|
6 |
28 |
Graduate Division NRSA Training Grants Teaching Assistantships Extramural Research Grants |
Section 8. Changes in Senate Regulations
None.
Section 9. Abstract
Section 10.
Departmental Commitment to Proposed Program
See Appendix D for
departmental letters of support for the proposed IDP.
Appendix A
Summary of
Information Required by
the California
Postsecondary Education Commission
1.
Name of Program: Bioinformatics Interdepartmental Degree
Program
2.
Campus: UCLA
3.
Degree/Certificate: M.S., Ph.D.
(The certificate
category referred to includes all organized programs which award certificates
for academic achievement. This includes professional certificate programs.
Skills programs which are designed specifically for state licensing purposes
need not be reported.)
4.
CIP Classification (to be completed by Office of the President):
5.
Date to be started: Fall Quarter 2007
6. Modification of
existing program, identify that program and explain changes. (This means new
programs that have roots in existing programs--which may or may not be degree
programs.) This is a new program.
7. Purpose (academic
or professional training), distinctive features and justification. How does
this program differ from others, if any offered in California. The document
issued by CPEC titled "Inventory of Academic and Occupational Programs in
California Colleges and Universities" is in the Graduate Council Office (extension
51162). The new program must be compared with similar programs listed in the
inventory.
UCLA has already established
a strong record of bioinformatics research and graduate training. In 1999 the faculty established a
graduate core curriculum in bioinformatics, which has been offered continuously
since that time, demonstrating the faculty’s commitment to collaborative
teaching and to long-term development of an integrated bioinformatics
program. These initiatives have
been recognized by a large number of awards of multi-investigator Project and
Training grants in bioinformatics from NIH, NSF, DOE and other funding
sources. These many disparate
efforts need a strong graduate program to make them cohesive, successful, and
competitive in the long term.
The establishment of the
Bioinformatics IDP will allow UCLA to overcome the limitations of the current
situation, in which no single program brings together bioinformatics
students. Specifically, we expect
to resolve these existing weaknesses:
- Prospective bioinformatics graduate students do
not know which program to apply to at UCLA, since neither departmental
graduate programs or ACCESS specifically recruit and train them.
- Graduate students at UCLA who are conducting
bioinformatics research currently take diverse courses that do not
necessarily cover the core training in bioinformatics that we believe they
need and that we propose here.
- Relative to other UC schools that already have
established bioinformatics graduate programs, the lack of such a program
at UCLA places UCLA at a disadvantage in competing for the top
bioinformatics students, and this impacts the ability of our faculty to
obtain funding in this area.
Comparisons
with specific programs at other universities are detailed under item 14 below
and in the proposal.
8.
Type(s) of students to be served. Graduate students
seeking integrated training in bioinformatics in an interdisciplinary setting.
9. If program is not
in current campus academic plan, give reason for starting program now.
In 1999 the School of
Medicine conducted a strategic planning process, which identified
bioinformatics as one of the highest priorities for immediate development. One of the recommendations to emerge
from this review was for the deans to develop a specific plan for
bioinformatics training, which included a degree-granting education
program. The fact that this
recommendation was made five years ago indicates that the need for the proposed
IDP is urgent. It is essential
because it would bring together UCLA’s many academic resources in
bioinformatics for the first time.
The interdisciplinary character of UCLA’s bioinformatics
faculty—scattered among 11 departments in four schools and
divisions—is both a strength and a potential weakness. As other prominent universities develop
bioinformatics graduate degree programs and competition grows, UCLA will be at
a severe disadvantage in recruiting top graduate student candidates if it does
not offer a bioinformatics degree and admissions program. Failure to establish a graduate program
would have negative effects on all of the existing initiatives in
bioinformatics and genomics at UCLA.
Long-term, it is hard to imagine that the bioinformatics training grant
awards that UCLA has received would be renewed if the campus failed even to
begin a graduate program. Faculty
research in bioinformatics depends critically upon the quality of graduate
student recruitment. Lack of a
bioinformatics graduate program would become a major disadvantage once other
universities establish strong bioinformatics degree programs, which is already
happening. This in turn would hurt
the quality of bioinformatics research at UCLA, and make it difficult to retain
top bioinformatics faculty or recruit new faculty.
10. If program
requires approval of a licensure board, what is the status of such approval? Not applicable.
11. Please list
distinctive features of the program having the character of credit for
experience, internships, lab requirements, etc. Not
applicable.
12.
List all new courses required:
|
Department |
Course Number |
Title |
Hours/Week Lecture
& Lab |
|
Chemistry |
260B |
Advanced
Algorithms for Bioinformatics |
4
lecture, 4 lab |
|
Chemistry |
260BL |
Bioinformatics
Algorithms Laboratory |
4
lab |
13.
List all other required courses:
|
Department |
Course Number |
Title |
Hours/Week Lecture/
Discussion/Lab |
Notes |
|
Chemistry |
260A |
Bioinformatics
and Genomics |
3
lecture, 1 discussion |
formerly
Chemistry 260 |
|
Chemistry |
202 |
Bioinformatics
Interdisciplinary Research Laboratory |
2
lecture, 2 discussion |
|
|
Stats |
254 |
Statistical
Methods in Computational Biology |
3
lecture, 1 discussion |
to
be listed also as Chem M260C in future |
14. List UC campus
and other California institutions, public or private, which now offer or plan
to offer this program or closely related programs. (The current requirement is
that these programs be listed. What is of concern is possible duplication.
Proposal sponsors should be aware of this and give careful attention to the
program justification in #6.)
|
University |
Program Name |
Degrees Granted |
Notes |
|
Stanford
University |
Biomedical
Informatics |
Ph.D. |
focused
on biomedical applications |
|
UC
Berkeley |
Computational
and Genomic Biology |
Designated
Emphasis in existing Ph.D. programs |
a
minor only |
|
UC
Riverside |
Genetics,
Genomics and Bioinformatics |
Ph.D. |
bioinformatics
part of GGB program, not main focus |
|
UC
San Francisco |
Biological
and Medical Informatics |
M.S.,
Ph.D. |
based
in medical informatics |
|
UC
Santa Cruz |
Bioinformatics |
B.S.,
M.S., Ph.D. |
not
interdepartmental; less breadth in core faculty |
|
UC
San Diego |
Bioinformatics |
Ph.D.;
also a graduate specialization |
closest
to proposed program |
|
University
of Southern California |
Computational
Biology and Bioinformatics |
Ph.D. |
not
interdepartmental |
15.
List any related program offered by the proposing institution and explain
relationship.
ACCESS: umbrella graduate admissions program,
does not include many bioinformatics faculty, no bioinformatics component in
training
Biomathematics: does not involve bioinformatics
requirements or research
Biomedical
Engineering: does not require
bioinformatics, and most bioinformatics faculty are outside of the School of
Engineering
Human
Genetics: requirements focused on
human genetics; most bioinformatics faculty are not in this department
16.
Summarize employment prospects for graduates of the proposed program. Give
results of job market survey if such have been made. (This is aimed at graduate
or undergraduate professional programs.)
Bioinformatics is of central
importance to biomedical research in the 21st century and to the
economy of California, and employment prospects are expected to be
excellent. To demonstrate the
strong market demand for bioinformaticists the proposal lists the job openings
posted on the web site of the International Society for Computational Biology
for July through September of 2006 --
a total of about 50 listings in both academia and industry.
17.
Give estimated new and total enrollment for the first five years and state
basis for estimate.
Projected Doctoral Enrollments
|
Year |
New |
Continuing |
Total |
|
1 |
3 |
0 |
3 |
|
2 |
5 |
3 |
8 |
|
3 |
5 |
8 |
13 |
|
4 |
5 |
13 |
18 |
|
5 |
5 |
18 |
23 |
18.
Give estimates of the additional cost of the program by year for five years in
each of the following categories:
The
development of this degree program utilized resources already available to
faculty and
research
programs without significant additional costs.
FTE Faculty: New
faculty in bioinformatics may be hired by specific departments in future, but
this proposal does not require this.
Library Acquisitions: The Library’s current holdings are sufficient, assuming
appropriate updates.
Computing: Existing
computing facilities will be utilized.
Other Facilities:
None.
Equipment: Existing
equipment will be utilized.
Provide brief explanation of any of the costs where
necessary.
(The
additional resources, if any, required by the proposed program must be included
even if they can be supplied by campus reallocation.)
No
additional costs are anticipated.
19. How and by what
agencies will the program be evaluated? (This refers to the campus and
professional review procedures.)
The IDP will first be reviewed after two years of operation,
by an internal review committee consisting of the Executive Committee and
additional members appointed by the Deans of the Schools of Medicine and
Engineering and the Deans of the Divisions of Life Sciences and Physical
Sciences. Results of the review
will be provided to all four Deans.
Subsequently, the IDP will undergo external review every eight
years. We will also establish an
external advisory board consisting of distinguished scientists from industry.
Appendix B
Letter of Support
from Comparable California Programs
Appendix C
Brief Curricula Vitae
of Program Faculty
Appendix D
Letters of Support
from UCLA Departments
Appendix E
Letters of Interest from Participating Faculty Members
Appendix F
Catalog Descriptions
of all Required and Recommended Courses
(Current as of
1/25/07)
Required Courses:
Chemistry 202.
Bioinformatics Interdisciplinary Research Seminar. (4)
Seminar, two hours;
discussion, two hours. Concrete examples of how biological questions about
genomics data map to and are solved by methodologies from other disciplines,
including statistics, computer science, and mathematics. May be repeated for
credit. S/U or letter grading.
Chemistry CM260A.
Introduction to Bioinformatics and Genomics. (4)
(Formerly numbered CM260.)
(Same as Human Genetics M260A.) Lecture, three hours; discussion, one hour.
Recommended requisite: Statistics 100A or 110A. Genomics and bioinformatics
results and methodologies, with emphasis on concepts behind rapid development
of these fields. Focus on how to think genomically via case studies showing how
genomics questions map to computational problems and their solutions.
Concurrently scheduled with course C160A. S/U or letter grading.
Chemistry C260B. Algorithms in Bioinformatics and
Systems. (4)
Lecture, 4 hours; laboratory,
4 hours. Enforced requisite: C160A or C260A with a grade of C- or
better. Recommended: Statistics 100A and 110A and PIC 32 and
60. Development and
application of computational approaches to biological questions. Understanding
of mechanisms for determining statistical significance of computationally
derived results. Students will develop a foundation for innovative work in
Bioinformatics and Systems Biology.
Concurrently scheduled with course C160B. S/U or letter grading.
Chemistry 260BL. Advanced Bioinformatics Computational
Laboratory. (2)
Laboratory, 4 hours. Enforced requisite: CM260A. Co-requisite: C260B. This course will focus on the
development and application of computational approaches to ask and answer
biological questions. Students completing the course should be able to
implement a variety of bioinformatics and systems biology algorithms.
Correspondingly, they should have an appreciation for the advantages and
disadvantages of different algorithmic methods for studying biological
questions. Furthermore, students should gain a preliminary understanding of how
to compute the statistical significance of their results, a process which may
involve writing an estimation or sampling program. The course will focus on
development of a conceptual understanding of implementation of bioinformatics
algorithms and give students a foundation for how to do innovative work in
these fields. Material will be drawn from specific, relevant biological
problems and will closely parallel 260B. As a complement to 260, students will
gain experience in observing the impact of computational complexity of an
algorithm in computing a solution.
S/U or letter grading.
Statistics M254.
Statistical Methods in Computational Biology. (4)
(Same as Biomathematics
M271.) Lecture, three hours; discussion, one hour. Preparation: elementary
probability concepts. Requisite: course 100A. Training in probability and
statistics for students interested in pursuing research in computational
biology, genomics, and bioinformatics. Letter grading. (To be multiple-listed
in future as Chemistry 260C.)
Elective Courses:
Biomathematics M203. Stochastic Models in Biology. (4)
(Same as Human Genetics
M203.) Lecture, four hours. Requisite: Mathematics 170A or equivalent
experience in probability. Mathematical description of biological
relationships, with particular attention to areas where conditions for
deterministic models are inadequate. Examples of stochastic models from
genetics, physiology, ecology, and a variety of other biological and medical
disciplines. S/U or letter grading.
Biomathematics M207A. Theoretical Genetic Modeling. (4)
(Same as Biostatistics M272
and Human Genetics M207A.) Lecture, three hours; discussion, one hour.
Requisites: Mathematics 115A, 131A, Statistics 100B. Mathematical models in
statistical genetics. Topics include population genetics, genetic epidemiology,
gene mapping, design of genetics experiments, DNA sequence analysis, and
molecular phylogeny. S/U or letter grading.
Biomathematics 210.
Optimization Methods in Biology. (4)
Lecture, four hours.
Preparation: undergraduate mathematical analysis and linear algebra;
familiarity with programming language such as Fortran or C. Modern
computational biology relies heavily on finite-dimensional optimization. Survey
of theory and numerical methods for discrete and continuous optimization, with
applications from genetics, medical imaging, pharmacokinetics, and statistics.
S/U or letter grading.
Biomathematics M211. Mathematical and Statistical Phylogenetics.
(4)
(Same as Human Genetics
M211.) Lecture, three hours; laboratory, one hour. Requisites: Biostatistics
110A, 110B, Mathematics 170A. Theoretical models in molecular evolution, with
focus on phylogenetic techniques. Topics include evolutionary tree
reconstruction methods, studies of viral evolution, phylogeography, and
coalescent approaches. Examples from evolutionary biology and medicine.
Laboratory for hands-on computer analysis of sequence data. S/U or letter
grading.
Biomedical Engineering M217.
Biomedical Imaging. (4)
(Same as Electrical
Engineering M217.) Lecture, three hours; laboratory, two hours; outside study,
seven hours. Requisite: Electrical Engineering 114D or 211A. Mathematical
principles of medical imaging modalities: X-ray, computed tomography,
positron-emission tomography, single photon emission computed tomography,
magnetic resonance imaging. Topics include basic principles of each imaging
system, image reconstruction algorithms, system configurations and their
effects on reconstruction algorithms, specialized imaging techniques for
specific applications such as flow imaging. Letter grading.
Biomedical Physics 208A.
Medical Physics Laboratory: Medical Imaging. (4)
Discussion, two hours;
laboratory, four hours. Requisite: course 205. Hands-on experience performing
acceptance testing and quality control checks of imaging equipment such as
fluoroscopy, digital subtraction angiography, mammography, ultrasound, magnetic
resonance imaging, computed tomography, and computed radiography.
Biomedical Physics 210.
Principles of Medical Image Processing. (4)
Lecture, three hours; discussion,
one hour. Requisite: course 209. Study of image representation,
computational structures for imaging, linear systems theory, image enhancement
and restoration, image compression, segmentation, and morphology. Special
topics include visualization techniques, three-dimensional modeling, computer
graphics, and neural net applications. Laboratory projects apply concepts
developed in class.
Biomedical Physics 214.
Medical Image Processing Systems. (4)
Lecture, three hours;
discussion, one hour. Requisites: courses 209, 210. Advanced image processing
and image analysis techniques applied to medical images. Discussion of
approaches to computer-aided diagnosis and image quantitation, as well as
application of pattern classification techniques (neural networks and
discriminant analysis). Examination of problems from several imaging modalities
(CT, MR, CR, and mammography).
Biomedical Physics M266. Advanced Magnetic Resonance Imaging. (4)
(Same as Neuroscience M267
and Psychiatry M266.) Lecture, four hours. Starting with basic principles,
presentation of physical basis of magnetic resonance imaging (MRI), with
emphasis on developing advanced applications in biomedical imaging, including
both structural and functional studies. Instruction more intuitive than mathematical.
Letter grading.
Biomedical Physics M285.
Functional Neuroimaging: Techniques and Applications. (4)
(Same as Psychiatry M285.)
In-depth examination of activation imaging, including PET and MRI methods, data
acquisition and analysis, experimental design, and results obtained thus far in
human systems. Strong focus on understanding technologies, how to design
activation imaging paradigms, and how to interpret results. Laboratory visits
and design and implementation of a functional MRI experiment. S/U or letter
grading.
Biostatistics M272.
Theoretical Genetic Modeling . (4)
(Formerly numbered M237A.)
(Same as Biomathematics M207A and Human Genetics M207A.) Lecture, three hours;
discussion, one hour. Requisites: Mathematics 115A, 131A, Statistics 100B.
Mathematical models in statistical genetics. Topics include population
genetics, genetic epidemiology, gene mapping, design of genetics experiments,
DNA sequence analysis, and molecular phylogeny. S/U or letter grading.
Biostatistics M278. M278. Statistical Analysis of DNA
Microarray Data. (4)
(Formerly numbered 278.)
(Same as Human Genetics M278.) Lecture, three hours. Requisite: course 200C.
Instruction in use of statistical tools used to analyze microarray data.
Structure corresponds to analytical protocol an investigator might follow when
working with microarray data. S/U or letter grading.
Chemistry 298. Seminar: Computational Methods in
Bioinformatics. (To be proposed.) Discussion, one hour. Detailed analysis
of new methods, including algorithms and representation of bioinformatics data,
and research problems ranging from genome-wide sequence analysis, to fold
recognition, to energy calculation and protein design.
Computer Science 249.
Current Topics in Data Structures. (2 to 12)
Lecture, four hours; outside
study, eight hours. Review of current literature in an area of data structures
in which instructor has developed special proficiency as a consequence of
research interests. Students report on selected topics. May be repeated for
credit with consent of instructor. Letter grading.
Ecology and Evolutionary
Biology M231. Molecular Evolution. (4)
(Same as Earth and Space
Sciences M217.) Lecture, two hours; discussion, two hours. Series of advanced
topics in molecular evolution, with special emphasis on molecular
phylogenetics. Topics may include nature of the genome, neutral evolution,
molecular clocks, concerted evolution, molecular systematics, statistical
tests, and phylogenetic algorithms. Themes may vary from year to year. May be
repeated for credit. S/U or letter grading.
Human Genetics C244.
Genomic Technology. (4)
Lecture, three hours;
discussion, one hour. Requisite: Life Sciences 4. Survey of key technologies
that have led to successful application of genomics to biology, with focus on
theory behind specific genome-wide technologies and their current applications.
Concurrently scheduled with course C144. S/U or letter grading.
MCDB 292. Seminar: Molecular Evolution. (2)
Discussion, three hours.
Detailed analysis of current understanding of evolution of molecular sequences
and structures.
Neuroscience CM272.
Neuroimaging and Brain Mapping. (4)
(Same as Physiological
Science M272 and Psychology M213.) Lecture, three hours. Requisites: courses
M201, M202. Theory, methods, applications, assumptions, and limitations of
neuroimaging. Techniques, biological questions, and results. Brain structure,
brain function, and their relationship discussed with regard to imaging.
Concurrently scheduled with course C172. Letter grading.
Pathology 255. Mapping and Mining Human Genomes. (3)
Lecture, three hours. Basic
molecular genetic and cytogenetic techniques of gene mapping. Selected regions
of human genomic map scrutinized in detail, particularly gene families and
clusters of genes that have remained linked from mouse to human. Discussion of
localizations of disease genes. S/U or letter grading.
Statistics 165.
Statistical Methods and Data Mining. (4)
Lecture, three hours.
Requisite: course 100A. Introduction and overview of up-to-date statistical
methods in microarray analysis designed for students in biostatistics,
statistics, and human genetics who are interested in technology and statistical
analysis of microarray experiments. Useful for biology students with basic
statistical training who are interested in understanding logic underlying many
statistical methods. P/NP or letter grading.
Statistics 233.
Statistical Methods in Biomedical Imaging. (4)
Lecture, three hours.
Requisite: course 100A. Brief review of common general statistical techniques.
Advanced statistical methods for analysis of medical imaging, integration,
visualization, interrogation, and interpretation of imaging and nonimaging
metadata. S/U or letter grading.
Appendix G
Degree Program
Requirements
Bioinformatics
Interdepartmental Program
Graduate Degrees
The Bioinformatics Program offers the Master of Science (M.S.) and Doctor of
Philosophy (Ph.D.) degrees in Bioinformatics.
Admission
|
Program Name |
Bioinformatics Bioinformatics is an interdepartmental
program. Interdepartmental programs provide an integrated curriculum
of several disciplines. |
|
Address |
xxx |
|
Phone |
(310) xxx-xxxx |
|
Email |
xxx@ucla.edu |
|
Leading to the degree of |
M.S., Ph.D. |
|
Admission Limited to |
Fall |
|
Deadline to apply |
December 1st |
|
GRE (General and/or Subject), TSE, TWE |
GRE: General and Subject in Biology, Biochemistry, Computer Science or
Mathematics. |
|
Letters of Recommendation |
3, from professors, supervisors, or others who may provide an evaluation
of the applicant's accomplishments or potential in research, teaching, and
related scholarly activities |
|
Other Requirements |
In addition to the University's minimum requirements and those listed
above, all applicants are expected to submit a statement of purpose. The department encourages applications from students in all areas of
science, but expects successful applicants to have or to acquire a background
comparable to the requirements for the bachelor's degree in Computational and
Systems Biology at UCLA. A background in chemistry, physics, and mathematics
is desirable. Deficiencies in these or other subjects should be made up at
the earliest opportunity. Undergraduates who are prospective applicants
should remedy their deficiencies by preparatory study at an appropriate
institution. Students with academic deficiencies may be admitted on a
provisional basis. |
Master's Degree
Advising
All academic affairs for graduate students in the department are directed by
the Interdepartmental Program Graduate Adviser who is assisted by the
administrative staff of the Graduate Affairs Office. The Graduate Adviser
establishes, at the time of admission to graduate study, a guidance committee
for each student that consists of three faculty members for each student.
The chair of the guidance committee acts as the provisional adviser until a
permanent adviser is selected. Service as a provisional adviser is designed to
be provisional for both professor and student. It does not commit the professor
to supervise the thesis, nor does it commit the student to a provisional
adviser. The provisional adviser serves until a permanent adviser is found and
the master's examination or thesis committee is established.
Areas of Study
Study consists of coursework and research within the program in a core
curriculum, genomics, proteomics, neuroinformatics, computer science, and
mathematics and statistics.
Foreign Language Requirement
None.
Course Requirements
The program consists of at least nine courses completed in graduate
standing, of which at least five must be graduate (200-series) courses. The
remaining courses may be in the 100, 200, or 500 series. No more than two 596
courses (eight units) may be applied toward the nine courses required for the
degree; only one 596 course (four units) may be applied toward the minimum five
graduate courses required. Courses in the major that are taken for S/U grading
may not be applied toward the minimum requirement. Courses outside of the major
that are taken for S/U grading may be applied toward the minimum requirement if
they are deemed applicable and provided that no more than one such course is
taken per quarter. Students must take the core curriculum courses (Chemistry
and Biochemistry 260A, Chemistry and Biochemistry 260B, Statistics 254,
Chemistry and Biochemistry 260BL, Chemistry and Biochemistry 202, and Chemistry
and Biochemistry 252), plus four elective courses from concentration areas, and
at least 12 units of research.
Teaching Experience
One quarter of teaching experience is required.
Field Experience
Not required.
Comprehensive Examination Plan
Students who select this plan are required to take an oral examination. The advisory committee evaluates
and grades the proposal as not pass or M.A. pass and forwards the results to
the Graduate Adviser.
Thesis Plan
A student must choose an IDP faculty advisor and submit a dissertation
proposal by the end of the third quarter of study. To proceed, the proposal must be approved by a faculty
member who agrees to become the student’s dissertation advisor. Completed dissertations will be
evaluated by a committee of at least three IDP faculty members, which must be
approved by the IDP Director. The
student must present the completed dissertation in a public seminar.
Time-to-Degree
The normative time-to-degree for the master's degree is five to six
quarters.
Doctoral Degree
Advising
The Bioinformatics program provides a comprehensive system of advising for
students throughout their graduate studies. During orientation the advising
committee and program chair meet with new students to review the first-year
requirements in general terms. Throughout the term, students are expected to
meet individually with the chair or other members of the advising committee to
identify faculty whose research is closest to their own interests and who would
be most appropriate for laboratory rotations. At the end of the fall term, the
entire advising committee meets informally with the first-year students to
field questions that have come up after their initial entry into the program. In
subsequent quarters, students' enrollment and performance in core courses and
laboratory rotations are closely monitored and, as the need arises, students
are counseled individually by the
advising chair. At the end of Spring Quarter of the first year, students are
required to submit a Faculty Mentor Approval Form (co-signed by the mentor) to
the advising committee, which meets to consider the choice of mentor and the
ability of the faculty to serve in this capacity.
The advising program continues after each student has chosen a faculty
research mentor. Every year students receive a memorandum outlining current
requirements (for example, course electives, the written and oral qualifying
examinations and midstream seminar). The advising committee also meets every
year to discuss the progress of all students and identify potential problems.
The committee then sends each student a letter that assesses their current
progress in the program and makes specific recommendations as needed. An
overall assessment of student progress is also made annually to the
neuroscience committee. In addition to the formal advising procedures outlined
above, students are repeatedly encouraged to seek advice on career development
from faculty members in the UCLA Bioinformatics community. Finally, an annual
retreat serves the purpose of allowing informal and organized contacts between
faculty and students, which provides further opportunity for advising.
Major Fields or Subdisciplines
These fields include genomics, proteomics, neuroinformatics, computer
science, and mathematics and statistics.
Foreign Language Requirement
None.
Course Requirements
Students must take the core curriculum courses (Chemistry and Biochemistry
260A, Chemistry and Biochemistry 260B, Statistics 254, Chemistry and
Biochemistry 260BL, Chemistry and Biochemistry 202, and Chemistry and
Biochemistry 252), plus five elective courses from concentration areas. They must also complete 12 units of
laboratory rotation and at least 6 units of seminar courses.
Teaching Experience
One quarter of teaching experience is required.
Written and Oral Qualifying Examinations
Academic Senate regulations require all doctoral students to complete and
pass University written and oral qualifying examinations prior to doctoral
advancement to candidacy. Also, under Senate regulations the University oral
qualifying examination is open only to the student and appointed members of the
doctoral committee. In addition to University requirements, some graduate
programs have other pre-candidacy examination requirements. What follows in
this section is how students are required to fulfill all of these requirements
for this doctoral program.
Ph.D. degree candidates, but not M.S. candidates, will be
required to pass a written qualifying examination, consisting of a research
proposal outside their dissertation research topic, and an oral qualifying exam
defending their dissertation research proposal before their dissertation
committee. Moreover, before taking
the Ph.D. qualifying exams, the student must first complete the Core
Curriculum. Following qualifying
exams and advancement to candidacy, students must present a summary of their
research to date and proposed future research within an oral seminar
format. This summary should be
presented approximately one year after advancement to candidacy.
During their first year, Ph.D. students will perform
laboratory rotations with IDP faculty whose research interests them, and must
select a dissertation advisor from the IDP faculty by the end of their third
quarter. Next, students must
select a qualifying committee by the end of their second Spring quarter, and
submit a written dissertation proposal by the end of their second year. The qualifying committee must consist
of the faculty advisor and at least three additional faculty, of whom two must
be IDP faculty. The qualifying
committee must be approved by the IDP Director and the campus graduate
Dean. The written dissertation
proposal must be given to all the qualifying committee members prior to the
scheduled oral qualifying exam.
Advancement to Candidacy
Students will advance to candidacy when they pass the oral qualifying exam,
complete the IDP course requirements, clear all incompletes from their
transcript, and pay the filing fee.
Students should advance to candidacy by the end of their third Fall
quarter. Failure to advance to
candidacy by the end of the fourth year will result in academic probation.
Doctoral Dissertation
Every doctoral degree program requires the completion of an approved dissertation
that demonstrates the student's ability to perform original, independent
research and constitutes a distinct contribution to knowledge in the principal
field of study.
The completed dissertation must be submitted to the reading
committee three weeks before the dissertation defense. The reading committee must consist of
the faculty advisor and at least two additional readers from the IDP faculty,
and must be approved by the IDP Director.
The candidate must present a public seminar, immediately followed by the
dissertation defense. The
committee must determine whether the dissertation completely fulfills the
requirements of original, important, publishable research in bioinformatics,
and whether revisions are required.
Ph.D. dissertations must conform to a high standard of originality,
relevance and impact suitable for publication in peer-reviewed journals in the
field. Successful completion of
the dissertation and defense constitutes the final requirement for the Ph.D.
degree.
Final Oral Examination (Defense of Dissertation)
Required for all students in the program.
Time-to-Degree
In general, overall progress toward the degree is accomplished with
completion of the written qualifying examination by the beginning of the second
year. It is recommended that students complete the University Oral Qualifying
Examination by the end of Spring Quarter of the third year. The approved normative time-to-degree
is 16 quarters.
Termination of Graduate Study and Appeal of Termination
University Policy
A student who fails to meet the above requirements may be recommended for
termination of graduate study. A graduate student may be disqualified from
continuing in the graduate program for a variety of reasons. The most common is
failure to maintain the minimum cumulative grade point average (3.00) required
by the Academic Senate to remain in good standing (some programs require a
higher grade point average). Other examples include failure of examinations,
lack of timely progress toward the degree and poor performance in core courses.
Probationary students (those with cumulative grade point averages below 3.00)
are subject to immediate dismissal upon the recommendation of their department.
University guidelines governing termination of graduate students, including the
appeal procedure, are outlined in Standards and Procedures for Graduate
Study at UCLA.
Special Departmental or Program Policy
A student must receive at least a B- in each core course or repeat the
course. A student who receives three B- grades in the core courses, who
fails all or part of the written or oral qualifying examinations two times
(if the student fails all or part of the written qualifying examination the
examination committee determines the form of reexamination), or who fails to
maintain minimum progress may be recommended for termination by vote of the
entire interdepartmental degree committee. A student may appeal a
recommendation for termination in writing to the interdepartmental degree
committee and may personally present additional or mitigating information to
the committee, in person or in writing.
Appendix I
Existing
Bioinformatics Equipment Resources